1. Introduction
The electrocardiogram (ECG) stands as a cornerstone in the diagnostic toolkit for cardiovascular diseases, a leading global cause of mortality (Collaborators GBDCoD, 2018). As ECG technology has evolved from analog to digital formats, automated computer analysis has become increasingly prevalent and successful in medical diagnoses (Willems et al., 1987; Schlapfer and Wellens, 2017). Deep learning methodologies, in particular, have demonstrated exceptional diagnostic capabilities in classifying ECGs using signal data, in some instances even outperforming individual cardiologists. For instance, a study leveraging raw ECG data developed a deep neural network (DNN) that matched or exceeded the average cardiologist’s accuracy in classifying 12 distinct cardiac rhythms in single-lead ECGs, including conditions like atrial fibrillation/flutter, atrioventricular block, junctional rhythm, and supra/ventricular tachycardia (Hannun et al., 2019). Further research utilizing signal data from 12-lead ECGs has also yielded impressive outcomes in arrhythmia classification (Baek et al., 2021).
While the promise of automated ECG diagnosis to enhance clinical workflows is significant, many existing models are designed to diagnose singular clinical pathologies. This approach limits their utility as ECGs frequently present with multiple concurrent abnormalities (Biton et al., 2021; Raghunath et al., 2021). Moreover, a majority of these diagnostic tools rely on the analysis of raw signal data (Hannun et al., 2019; Hughes et al., 2021; Sangha et al., 2022). This reliance poses a considerable challenge in clinical settings where ECGs are often printed on paper or accessible only as digital images. This is especially pertinent in remote and regional areas, often characterized by a scarcity of specialist medical expertise (Schopfer, 2021). In such environments, image-based deep learning models for ECG recognition would be invaluable, yet research in this area remains limited. A prior study explored an image-based model to differentiate between normal and abnormal ECGs, while another achieved high accuracy (99.05%) and sensitivity (97.85%) for 7 cardiac conditions through analysis of individual ECG beats (Jun et al., 2018). More recently, a model surpassing signal-based imaging in performance achieved an area under the receiver operating characteristic curve (AUROC) of 0.99 and an area under the Precision-Recall curve (AUPRC) of 0.86 for 6 clinical disorders (Sangha et al., 2022).
To our knowledge, this research represents a pioneering effort in training a convolutional neural network (CNN) to classify raw images of 12-lead ECGs for 10 distinct pathologies. Our methodology distinguishes itself from most prior studies by directly employing ECG image data for training and testing deep learning models, rather than relying on raw signal data or its transformations. This image-based approach directly addresses the practical limitations encountered in resource-constrained clinical environments, making advanced diagnostic tools more accessible and applicable.
2. Materials and methods
2.1. Datasets
The primary dataset for model development and evaluation was PTB-XL, a large, publicly available electrocardiography dataset. To assess the external validity of our models, we also conducted tests using unseen datasets, both individually and in combination.
The following publicly accessible ECG datasets were utilized in this study:
- PTB-XL (PTB) (Wagner et al., 2020): A comprehensive dataset for electrocardiography research.
- CPSC 2018 database (CPSC) (Liu et al., 2018): A database from the China Physiological Signal Challenge focused on ECG rhythm and morphology abnormality detection.
- 12-lead ECG database for arrhythmia research from Chapman University and Shaoxing People’s Hospital (Shaoxing) (Zheng et al., 2020): A dataset designed for arrhythmia research, encompassing over 10,000 patients.
- Test dataset for: Automatic multi-label ECG diagnosis of impulse or conduction abnormalities in patients with deep learning algorithm: a cohort study (Tongji) (Zhu et al., 2020): A dataset used to evaluate deep learning algorithms for automatic ECG diagnosis of heart rhythm and conduction abnormalities.
Each dataset provided raw 10-second 12-lead ECG signal waveform data alongside corresponding diagnostic labels, allowing for the generation of ECG images and the training of diagnostic models.
2.2. Data pre-processing
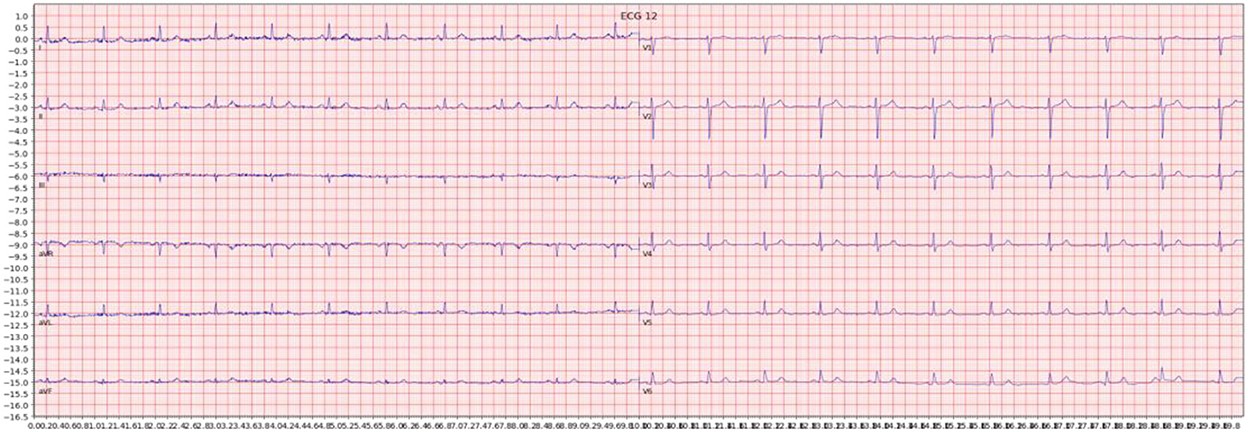
For each ECG sample, we generated an image by plotting the signal data using the Python ECG plot library (dy1901, 2022). This library produces 12-lead ECG images that closely resemble the displays and printouts commonly encountered in clinical practice. An example of such an image is shown below in Figure 1. All images were processed at a high resolution of 1600 × 512 pixels to maintain detail and clarity for the deep learning models.
Figure 1. Example of an ECG image generated from the signal data of one sample in the dataset.
The generated images then underwent a series of transformations to enhance feature extraction and model robustness. First, images were converted to grayscale to reduce color information and focus on waveform morphology. Subsequently, binarization was performed using three different techniques: simple thresholding, Otsu thresholding (an adaptive thresholding method), and adaptive thresholding. Each technique yields a binarized copy of the grayscale image, emphasizing different aspects of the ECG waveforms. To further augment the dataset and improve model generalization, a slightly blurred version of each grayscale image was created, and the same three thresholding techniques were applied to these blurred images. In total, eight augmented copies were generated from each original ECG image, expanding the dataset’s diversity and challenging the models to learn robust features. Figure 2 illustrates an example of an ECG image after grayscale conversion and adaptive thresholding.
Figure 2. Example of an ECG image after grayscale conversion and adaptive thresholding.
2.3. Classification task overview
Binary classification models were trained to predict the presence or absence of ten distinct cardiac conditions, covering a range of common and critical diagnoses in 12-lead ECG interpretation. These conditions were:
- Normal ECG (NORM)
- Left bundle branch block (LBBB)
- Right bundle branch block (RBBB)
- Atrial fibrillation (AFIB)
- Atrial flutter (AFLT)
- First-degree AV block (fAVB)
- Myocardial infarction (MI)
- Wolff-Parkinson-White syndrome (WPW)
- Supraventricular tachycardia
- Gender classification (hidden characteristic detection)
Models were primarily trained using the PTB-XL dataset and initially evaluated on a holdout test dataset from PTB-XL to assess internal validity. To evaluate the generalizability of the models, further testing was conducted using ECG images from external datasets not included in the training process. Additionally, tests were performed on combined datasets where matching diagnostic labels were available, to simulate more diverse clinical data scenarios. The results of these tests are summarized in Tables 1 and 2.
2.4. Model architecture and training
Our deep learning models were constructed using the VGG16 architecture, a well-established CNN framework known for its strong performance in image recognition tasks. We utilized ImageNet pre-trained weights for the VGG16 base, leveraging transfer learning to accelerate training and improve model performance. The original classification layer of the VGG16 model was removed and replaced with a custom classification head tailored to our 12-lead ECG diagnosis task. This head consisted of a global average pooling 2D layer to reduce spatial dimensions, a dropout layer for regularization during training, and a fully connected layer with a single output and sigmoid activation function for binary classification.
The training process was divided into two phases. Initially, only the newly added classification head was trained for up to 10 epochs with early stopping, while the VGG16 base layers remained frozen. This initial phase allowed the classification head to adapt to the ECG image data without disrupting the pre-trained feature extractors. Subsequently, the entire model, including the VGG16 base layers, was unfrozen and trained until no further reduction in validation loss was observed, again using early stopping with a patience of 6 epochs. A learning rate of 1 × 10-5 was used for the full model training. We also experimented with a learning rate schedule that reduced the learning rate upon validation loss plateauing, but this did not yield significant performance improvements. Binary cross-entropy loss was employed for most training instances, suitable for binary classification tasks. For datasets with significant class imbalance, we also explored the use of focal loss to address the imbalance issue, although binary cross-entropy proved sufficient for the majority of our experiments.
3. Results
The deep learning models developed in this study demonstrated robust performance when evaluated on unseen holdout test data derived from the same datasets used for training. This indicates strong internal validity and suggests the models effectively learned to recognize patterns within the training data distribution. However, generalization to completely unseen, external datasets proved to be more challenging, with performance generally lower than on the holdout test data. Interestingly, models trained on a combination of different datasets exhibited good performance on holdout test splits containing mixed data from those combined datasets. This suggests that training on diverse datasets can improve model robustness and generalization capacity. The detailed performance metrics, including AUROC, AUPRC, sensitivity, and specificity, are summarized in Tables 1 and 2.
3.1. Visual explanation
To enhance the interpretability of our deep learning models and gain insights into their decision-making processes, we employed Gradient-weighted Class Activation Mapping (Grad-CAM). Grad-CAM generates a heatmap that visually highlights the regions of an input image that are most influential in the model’s prediction for a given class. This technique allows us to examine which parts of the ECG images the model focuses on when making a diagnosis. Several illustrative examples of Grad-CAM heatmaps are presented below. Figure 3 demonstrates the heatmap highlighting delta waves in an ECG diagnosed with Wolff-Parkinson-White (WPW) syndrome. Figure 4 shows the heatmap emphasizing ST-segment changes in an ECG indicating myocardial infarction (MI). Figure 5 illustrates the heatmap focusing on the deep, broad S waves in lead V1, a characteristic feature of left bundle branch block (LBBB).
Figure 3. Example of an activation heatmap for an ECG showing WPW.
Figure 4. Example of an activation heatmap for an ECG showing myocardial infarction.
Figure 5. Example of an activation heatmap for an ECG showing LBBB.
4. Discussion
Our findings demonstrate that image-based deep learning models can achieve strong diagnostic performance on 12-lead ECGs, particularly when tested on data from populations and datasets similar to those used in training. This high level of internal validity is consistent with previous research on computer vision-based ECG models (Mohamed et al., 2015; Jun et al., 2018; Sangha et al., 2022). Jun et al. utilized a two-dimensional CNN to analyze individual ECG beats for detecting 7 distinct cardiac arrhythmias, while Sangha et al. developed a model based on the EfficientNet B3 architecture for examining 6 disorders, achieving performance comparable to or exceeding signal-based methods. In contrast, our model addressed a broader spectrum of up to 13 diagnoses, was trained on combinations of datasets, and underwent more extensive external validity testing. Table 3 provides a comparative overview, showing that the performance of our image-based model is comparable to other recent image-based studies as well as high-performing 12-lead ECG signal-based models. Notably, some of the most accurate signal-based models focused on single pathologies, such as atrial fibrillation, exhibit comparable findings to our study (e.g., AUROC of 0.997 vs. 1 and Sensitivity of 0.985 to 0.992 in Jo et al., 2021). While achieving comparable accuracy, our study advances beyond several signal-based studies in the literature that are limited by focusing on individual pathologies or single leads, thereby restricting their applicability to comprehensive 12-lead ECG readings and cases with multiple co-existing pathologies (Javadi et al., 2013; Biton et al., 2021; Raghunath et al., 2021).
A significant advantage of our image-based approach lies in its potential applicability in resource-limited settings. Current deep learning tools predominantly rely on signal data, which is not optimized for environments with limited resources, such as rural and remote healthcare facilities. A large proportion of ECGs in routine clinical practice are still either printed or scanned as images, which inherently restricts the utility of signal-based models in these contexts. Furthermore, while numerous models have been designed to accurately detect individual disorders, ECGs presenting with multiple co-existing abnormalities pose a considerable diagnostic challenge. Our model also demonstrated consistent and even superior performance compared to prior image-based studies in identifying patient gender from ECGs, both within internal and external datasets. This capability, also observed in signal-based models (Attia et al., 2019; Kim and Pyun, 2020), suggests that image-based models can accurately recognize subtle, “hidden” features within ECG data. This is particularly promising as ECG data is increasingly integrated with other patient observations and vital signs, which can be synergistically leveraged through advanced algorithms for enhanced clinical insights.
The incorporation of Gradient-weighted Class Activation Mapping (Grad-CAM) in our model provides crucial insights into the model’s decision-making process by highlighting image regions contributing to specific diagnostic labels. This feature enabled us to evaluate whether the assigned labels were based on clinically relevant information or potentially on spurious correlations within the data (DeGrave et al., 2020). We observed a strong correspondence between the features highlighted by Grad-CAM and those traditionally used by human experts in ECG interpretation. For example, Grad-CAM effectively highlighted delta waves in WPW (Figure 3), ST-segment changes in MI (Figure 4), deep broad S waves in V1 for LBBB (Figure 5), and prolonged PR segments in first-degree AV block (Figure 6), and QRS complexes without preceding P waves in atrial fibrillation. In a few instances, the highlighted regions were less directly relatable to human diagnostic criteria, such as emphasizing the area following an ectopic beat rather than the abnormally large QRS complexes themselves. These less intuitive activations occurred in a small percentage of cases and may be improved through further model training on more diverse datasets or by integrating advanced visualization technologies like HiResCAM (Draelos and Carin, 2020). In clinical application, presenting a Grad-CAM heatmap alongside the algorithmic diagnosis can provide valuable context and visual evidence supporting the diagnosis. This explainability can empower clinicians to make more informed decisions regarding the algorithm’s output, potentially facilitating smoother integration of AI-driven diagnostics into routine clinical practice (Makimoto et al., 2020).
Figure 6. Example of an activation heatmap for an ECG showing First Degree AV Block.
Regarding computational demands, our study utilized a PC equipped with a Ryzen 9 5900x CPU, RTX 3080 and 3080 Ti GPUs, and 64 GB RAM running on Linux Mint. Training times for fine-tuning the VGG16 model for binary classification of each diagnostic label ranged from 18 to 36 hours, concluding with early stopping based on validation AUROC plateauing. Inference times were rapid, in the range of 3–5 seconds per diagnosis label for each ECG image. While model training is computationally intensive, the swift inference times underscore the potential for real-time clinical application. Prior research has demonstrated that computerized ECG interpretations can significantly reduce analysis times (Schlapfer and Wellens, 2017), and our findings further highlight the potential of integrating computer vision algorithms into routine clinical care as a valuable adjunct for diagnosis and decision-making.
4.1. Future work and limitations
A notable limitation of our study is the observed decrease in model accuracy when applied to unseen external datasets. This performance decrement may stem from variations in diagnostic labeling criteria across different datasets or inconsistencies in ECG quality. Nonetheless, certain diagnostic labels, such as LBBB, RBBB, and WPW, were accurately classified even in external datasets (Table 4), indicating the potential utility of this technology on unseen data, provided sufficient and consistently labeled training data are available. Future research directions to enhance model generalization include incorporating more extensive pre-processing steps, such as randomly shuffling the positions of different leads on the ECG image to encourage the model to learn lead-invariant features. Furthermore, the inclusion of even more diverse and independent datasets in training is crucial. Exploring the integration of additional clinical information, such as patient age, gender, weight, and height, which are typically available to clinicians during ECG interpretation, could also bolster disease stratification models when combined with image-based algorithms. Finally, investigating hybrid architectures that combine the strengths of different deep learning models may yield superior results in future iterations.
5. Conclusion
This research provides compelling evidence that computer vision AI models can effectively diagnose cardiac conditions from 12-lead ECG images with promising accuracy. Future research aimed at translating this technology into clinical practice should prioritize developing models that generalize robustly across diverse ECG image formats and sources and encompass a broader range of clinically relevant diagnoses. Additional diagnoses of significant clinical interest include the detection of STEMI in patients with LBBB or pacemakers, differentiating supraventricular tachycardia with aberrancy from ventricular tachycardia, and classifying specific subtypes of AV block. Furthermore, future models should be designed to be adaptable to different ECG formats and presentation styles. The techniques demonstrated in this study also open avenues for novel practical applications, such as smartphone-based applications for diagnosing ECGs captured in photographs, or in telehealth settings where image-based ECG transmission is more feasible. Overall, the classification performance achieved on ECG images using deep CNNs is comparable to state-of-the-art models utilizing raw ECG signal holdout test data from the same datasets, highlighting the potential of image-based deep learning as a valuable tool in 12 Lead Ecg Diagnosis.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author contributions
RA was involved in data processing, training, and evaluating machine learning models. GH was involved in literature review and editing. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Attia, Z. I., Friedman, P. A., Noseworthy, P. A., Lopez-Jimenez, F., Ladewig, D. J., Satam, G., et al. (2019). Age and sex estimation using artificial intelligence from standard 12-lead ECGs. Circ. Arrhythm. Electrophysiol. 12, e007284. doi: 10.1161/CIRCEP.119.007284
Baek, Y. S., Lee, S. C., Choi, W., and Kim, D. H. (2021). A new deep learning algorithm of 12-lead electrocardiogram for identifying atrial fibrillation during sinus rhythm. Sci. Rep. 11,12818. doi: 10.1038/s41598-021-92172-5
Biton, S., Gendelman, S., Ribeiro, A. H., Miana, G., Moreira, C., Ribeiro, A. L. P., et al. (2021). Atrial fibrillation risk prediction from the 12-lead electrocardiogram using digital biomarkers and deep representation learning. Eur. Heart J.-Digit. Health. 2, 576–585. doi: 10.1093/ehjdh/ztab071
Collaborators GBDCoD (2018). Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 392, 1736–1788. doi: 10.1016/S0140-6736(18)32203-7
DeGrave, A. J., Janizek, J. D., and Lee, S. I. (2020). AI for radiographic COVID-19 detection selects shortcuts over signal. medRxiv. doi: 10.1101/2020.09.13.20193565
Draelos, R., and Carin, L. (2020). Use HiResCAM instead of Grad-CAM for faithful explanations of convolutional neural networks. arXiv.
dy1901 (2022). ECG Plot. Python 3 Reference Manual, Scotts Valley, CA: CreateSpace.
Hannun, A. Y., Rajpurkar, P., Haghpanahi, M., Tison, G. H., Bourn, C., Turakhia, M. P., and Ng, A. Y. (2019). Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med. 25, 65–69. doi: 10.1038/s41591-018-0268-3
Hughes, J. W., Olgin, J. E., Avram, R., Abreau, S. A., Sittler, T., Radia, K., et al. (2021). Performance of a convolutional neural network and explainability technique for 12-lead electrocardiogram interpretation. JAMA Cardiol. 6, 1285–1295. doi: 10.1001/jamacardio.2021.2746
Javadi, M., Arani, S. A. A. A., Sajedin, A., and Ebrahimpour, R. (2013). Classification of ECG arrhythmia by a modular neural network based on mixture of experts and negatively correlated learning. Biomed. Signal Process. Control. 8, 289–296. doi: 10.1016/j.bspc.2012.10.005
Jo, Y-Y., Cho, Y., Lee, S. Y., Kwon, J-M., Kim, K-H., Jeon, K-H., et al. (2021). Explainable artificial intelligence to detect atrial fibrillation using electrocardiogram. Int. J. Cardiol. 328, 104–110. doi: 10.1016/j.ijcard.2020.11.053
Jun, T., Nguyen, H., Kang, D., Kim, D., Kim, D., Kim, H., et al. (2018). Ecg arrhythmia classification using a 2-d convolutional neural network. arXiv.
Kim, B. H., and Pyun, J. Y. (2020). ECG identification for personal authentication using LSTM-based deep recurrent neural networks. Sensors. 20, 3069. doi: 10.3390/s20113069
Liu, F., Liu, C., Zhao, L., Zhang, X., Wu, X., Xu, X., et al. (2018). An open access database for evaluating the algorithms of electrocardiogram rhythm and morphology abnormality detection. J. Med. Imaging & Health Infor. 8, 1368–1373. doi: 10.1166/jmihi.2018.2442
Makimoto, H., Höckmann, M., Lin, T., Glöckner, D., Gerguri, S., Clasen, L., et al. (2020). Performance of a convolutional neural network derived from an ECG database in recognizing myocardial infarction. Sci. Rep. 10, 8445. doi: 10.1038/s41598-020-65105-x
Mohamed, B., Issam, A., Mohamed, A., and Abdellatif, B. (2015). ECG image classification in real time based on the haar-like features and artificial neural networks. Procedia Comput. Sci. 73, 32–39. doi: 10.1016/j.procs.2015.12.045
Raghunath, S., Pfeifer, J. M., Ulloa-Cerna, A. E., Nemani, A., Carbonati, T., Jing, L., et al. (2021). Deep neural networks can predict new-onset atrial fibrillation from the 12-lead ECG and help identify those at risk of atrial fibrillation-related stroke. Circulation. 143, 1287–1298. doi: 10.1161/CIRCULATIONAHA.120.047829
Ribeiro, A. H., Ribeiro, M. H., Paixão, G. M. M., Oliveira, D. M., Gomes, P. R., Canazart, J. A., et al. (2020). Automatic diagnosis of the 12-lead ECG using a deep neural network. Nat. Commun. 11, 1760. doi: 10.1038/s41467-020-15432-4
Sangha, V., Mortazavi, B. J., Haimovich, A. D., Ribeiro, A. H., Brandt, C. A., Jacoby, D. L., et al. (2022). Automated multilabel diagnosis on electrocardiographic images and signals. Nat. Commun. 13, 1583. doi: 10.1038/s41467-022-29153-3
Schlapfer, J., and Wellens, H. J. (2017). Computer-interpreted electrocardiograms: benefits and limitations. J. Am. Coll. Cardiol. 70, 1183–1192. doi: 10.1016/j.jacc.2017.07.723
Schopfer, D. W. (2021). Rural health disparities in chronic heart disease. Prev. Med. 152(Pt 2), 106782. doi: 10.1016/j.ypmed.2021.106782
Wagner, P., Strodthoff, N., Bousseljot, R-D., Kreiseler, D., Lunze, F. I., Samek, W., et al. (2020). PTB-XL, a large publicly available electrocardiography dataset. Scientific Data. 7, 154. doi: 10.1038/s41597-020-0495-6
Willems, J. L., Abreu-Lima, C., Arnaud, P., van Bemmel, J. H., Brohet, C., Degani, R., et al. (1987). Testing the performance of ECG computer programs: the CSE diagnostic pilot study. J. Electrocardiol. 20(Suppl):73–77.
Zheng, J., Zhang, J., Danioko, S., Yao, H., Guo, H., and Rakovski, C. (2020). A 12-lead electrocardiogram database for arrhythmia research covering more than 10,000 patients. Scientific Data. 7, 48. doi: 10.1038/s41597-020-0386-x
Zhu, H., Cheng, C., Yin, H., Li, X., Zuo, P., Ding, J., et al. (2020). Automatic multilabel electrocardiogram diagnosis of heart rhythm or conduction abnormalities with deep learning: a cohort study. Lancet Digit. Health. 2, e348–e357. doi: 10.1016/S2589-7500(20)30107-2
Keywords: ECG, diagnosis, classification, deep learning, 12-lead ECG
Citation: Ao R and He G (2023) Image based deep learning in 12-lead ECG diagnosis. Front. Artif. Intell. 5:1087370. doi: 10.3389/frai.2022.1087370
Received: 02 November 2022; Accepted: 19 December 2022; Published: 09 January 2023.
Edited by:
Kathiravan Srinivasan, Vellore Institute of Technology, India
Reviewed by:
Deepa Jose, KCG College of Technology, India
Emrullah Acar, Batman University, Turkey
Copyright © 2023 Ao and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raymond Ao, cmF5bW9uZDUzODkyQGdtYWlsLmNvbQ==
†These authors share first authorship