Clostridioides difficile infection (CDI) remains a significant healthcare challenge, necessitating rapid and accurate diagnostic strategies. While traditional methods for CDI diagnosis exist, they often present limitations in speed, sensitivity, and standardization. This article delves into the complexities of CDI diagnosis, examining current methodologies and advocating for optimized diagnostic algorithms to improve patient outcomes.
Current Diagnostic Challenges in CDI
The conventional gold standard for diagnosing CDI, involving cell culture, cytotoxicity assays, and neutralization tests (7), faces practical hurdles in modern clinical settings. These methods demand specialized expertise and equipment, are time-intensive, lack standardized protocols, and are prone to false-positive results due to stool protease activity. Consequently, the need for improved and efficient screening methods is critical (8). The optimal diagnostic algorithm for CDI remains a subject of debate, with many laboratories relying on toxin detection assays.
Rapid and cost-effective toxin assays like enzyme-linked immunosorbent assays (ELISAs) and card tests, targeting toxins A and B, suffer from suboptimal sensitivity. Furthermore, the instability of these toxins, which can be degraded by temperature fluctuations and fecal proteases, increases the likelihood of false-negative results (9,10).
Insights from Clinical Studies
A previous study conducted in 2005 (11) investigated the prevalence of toxigenic C. difficile in stool samples from outpatients, inpatients, and hospital staff. Utilizing culture and latex agglutination tests alongside ELISA for toxin A and B detection, the study revealed CDI positivity in 15.5% of outpatients and 17.1% of inpatients based on ELISA results. No hospital staff tested positive. Comparing toxin ELISA results with culture-latex agglutination tests, the toxin A latex method showed a sensitivity of only 30.7%, contrasting sharply with the 100% sensitivity of toxin ELISA. This study highlighted the consistent presence of C. difficile toxin in both hospital and community-acquired cases, reinforcing hospitalization as a significant CDI risk factor, as evidenced by higher toxin ELISA positivity in hospitalized patients.
Advanced age (>65 years) also emerges as a crucial risk factor for CDI (9). The aforementioned study found that 66.7% of C. difficile-positive patients were over 55 years old. While literature often links beta-lactam antibiotics to CDI, this study did not find a statistically significant correlation with specific antibiotic families in diarrhea cases (p=0.582).
The Role of GDH Assays in Diagnostic Algorithms
Given the limited sensitivity of toxin ELISA and card tests, glutamate dehydrogenase (GDH) ELISAs are frequently incorporated into diagnostic algorithms. GDH assays offer enhanced sensitivity because GDH antigens are present in both toxigenic and non-toxigenic C. difficile strains, enabling detection of a broader range of variants. However, despite high sensitivity, GDH assays exhibit lower specificity (12), necessitating confirmatory testing after a positive GDH result.
Leading healthcare organizations like the Society for Healthcare Epidemiology of America and the Infectious Diseases Society of America advocate for testing only amorphous stool samples from patients with diarrhea suspected of CDI, excluding ileus cases. Asymptomatic patient screening is discouraged. While fecal culture remains valuable for epidemiological research, its time-consuming nature and need for specialized expertise in toxigenic strain identification render it impractical for routine clinical use. Toxin ELISAs, while rapid, are diagnostically suboptimal due to low sensitivity. Latex agglutination kits for GDH detection show moderate sensitivity (58–68%) and high specificity (94–98%), while ELISA-based GDH kits offer better performance with 85–95% sensitivity and 89–99% specificity. A two-step diagnostic algorithm utilizing GDH as an initial screen followed by a confirmatory test is a widely accepted strategy. A negative GDH assay typically indicates a C. difficile-negative result, while a positive GDH necessitates further confirmation. Polymerase chain reaction (PCR)-based tests, known for their speed, sensitivity, and specificity, present a promising diagnostic avenue, although further validation is needed before widespread clinical adoption (13).
Two-Step and Multi-Step Diagnostic Algorithms in Practice
Research by LaSala et al. (14) analyzing 114 stool samples demonstrated the limitations of stand-alone toxin tests. In their study, varying positivity rates were observed across GDH, PCR, and toxin-based ELISA tests, highlighting potential false-negative results with toxin tests alone. Fenner et al. (15) implemented a two-step C. difficile diagnostic algorithm on 1468 stool samples, using PCR to resolve discrepancies. They found 12.7% GDH-positive samples and 36.9% toxin A/B positive samples. Importantly, ten GDH-negative patients tested positive for toxins, and PCR detected a low bacterial burden in five GDH-negative patients. Culture sensitivity in this study was reported as low, despite 52.9% of GDH- and toxin-positive patients having a positive toxigenic culture.
Crobach et al. (6) reviewed 18 diagnostic methods from 1991–2008, including various ELISAs, GDH methods, and PCR. In 85% of studies, the cytotoxicity test was the reference standard. Applying a two-step algorithm in a hypothetical cohort of 10,000 patients, with a 5% CDI prevalence, revealed that while the vast majority were correctly identified (true negatives and positives), false negatives and positives still occurred. The recommendation from this review favors a high-sensitivity GDH assay as the initial screening test, followed by a high-specificity toxin test for confirmation, specifically in diarrhea cases where other enteropathogens are ruled out. Williamson et al. (16) compared GDH assay alone to a two-step GDH-PCR algorithm in 7106 stool samples. GDH assay alone detected C. difficile in 4.7% of samples, while the two-step algorithm increased detection to 9.9%. These findings underscore the value of high-sensitivity GDH tests as initial screening tools to minimize false negatives. Babady et al. (17) comparing PCR and GDH culture cytotoxicity on 560 stool samples found that combining GDH culture cytotoxicity and PCR achieved 100% sensitivity, surpassing GDH-positive culture cytotoxicity alone.
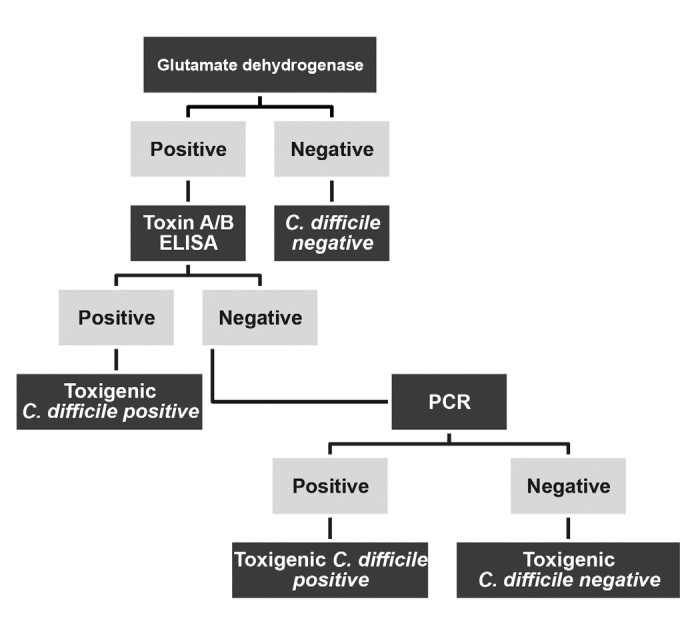 Diagnostic algorithm of Clostridium difficile
Diagnostic algorithm of Clostridium difficile
FIG. 1. Diagnostic algorithm of Clostridium difficile
Refining Diagnostic Accuracy and Algorithm Selection
Latex agglutination tests for toxin detection can produce false-positive results due to cross-reactions with both toxigenic and non-toxigenic C. difficile strains, as well as other bacteria like Peptostreptococcus anaerobius and Bacteroides asaccharolyticus (18). The observed inconsistencies between toxin-based ELISA and GDH methods in studies highlight PCR as a valuable confirmatory test for GDH-positive patients. GDH-positive, toxin-negative, PCR-positive results should be interpreted in conjunction with the patient’s clinical presentation, ruling out other causes of diarrhea before confirming a CDI diagnosis. However, it’s crucial to recognize that positive PCR results cannot differentiate between active CDI and asymptomatic colonization. A potential triple diagnostic algorithm incorporating GDH, toxin testing, and PCR is illustrated in Figure 1, offering a comprehensive approach to diagnosis.
| GDH/NAAT | Toxin ELISA | Comments |
|---|---|---|
| Positive | Positive | C. difficile Positive |
| Positive | Negative | Potential carriers of C. difficile |
| Negative | Negative | There is no C. difficile (Probably other pathogens) |
TABLE 4. Interpretation of C. difficile test results
GDH: glutamate dehydrogenase; NAAT: nucleic acid amplification testing
A survey of 170 hospitals in the United Kingdom (2009-2010) regarding C. difficile diagnostic algorithms (19) revealed that toxin A/B tests (ELISAs and card tests) were the most prevalent method (70%), while cell cytotoxicity neutralization tests were used by a small minority (3.6%). PCR was used as a primary screening method in only one laboratory. Notably, 19% of laboratories employed a two-step algorithm, and only 5% utilized a triple algorithm (GDH, toxin ELISA, and PCR), with another 5% using GDH and toxin A/B combination kits. This data underscores the variability in diagnostic practices and the potential for improvement through wider adoption of optimized algorithms.
Risk Factors, Test Performance, and Cost Considerations
The escalating prevalence and severity of CDI are linked to several key risk factors including prior antibiotic use, advanced age, healthcare facility stays, and possibly proton pump inhibitor use. Antibiotic use is implicated in most CDI cases, though sporadic cases occur even in healthy individuals without apparent risk factors (20). Fluoroquinolones and third-generation cephalosporins have been identified as frequently associated antibiotics in CDI patients with prior antibiotic exposure (21).
Accurate CDI diagnosis can be challenging despite available rapid tests due to varying test specificities (22). Stool samples in CDI cases might contain blood if colitis is severe, and fecal leukocytes are present in approximately half of cases. Stool assays for C. difficile, ranked from most to least sensitive, are: stool culture, GDH enzyme immunoassays (EIAs), RT-PCR, toxin A/B EIAs, and latex agglutination assays (23,24).
Stool culture, while highly sensitive, is labor-intensive and slow (48–96 hours), often yielding false positives due to non-toxigenic strains. However, it remains crucial for epidemiological studies. GDH EIAs offer high sensitivity and specificity (85–100% and 87–98%, respectively). Latex agglutination assays for GDH are less sensitive (48–59%) and specific (95–96%) compared to EIAs.
A study cohort demonstrated sensitivity, specificity, PPV, and NPV for toxin EIA as 67%, 94%, 21.4%, and 95.5%, and for GDH as 100%, 75%, 21.4%, and 100%, respectively. The low PPV of GDH alone highlights the necessity for additional testing of GDH-positive/toxin-negative specimens. Zheng et al. (26) reported good sensitivity but lower specificity and PPV for the Techlab C. diff Chek-60 GDH assay compared to cytotoxicity testing.
RT-PCR, detecting toxin genes, is highly specific and considered a potential gold standard alternative, albeit more expensive. Current gold standard cell culture toxin neutralization tests are less sensitive than PCR or toxigenic culture in diarrheal patients. Cost analysis reveals varying expenses for different tests, with toxin EIAs being among the most common screening methods due to high specificity, despite lower sensitivity (70–80%) compared to other methods.
Conclusion: Towards Optimized Diagnostic Algorithms for CDI
Many laboratories still use stand-alone toxin tests for CDI diagnosis, lacking standardization across healthcare facilities. This study reinforces the value of GDH EIA as an initial screening test, enhanced by a confirmatory toxin ELISA or card test to improve sensitivity and address potential false positives (27). PCR for toxigenic C. difficile offers superior sensitivity and specificity but its higher cost may limit widespread adoption. PCR’s ability to identify hypervirulent strains like 027/NAP-1 is epidemiologically significant. However, differentiation between ribotypes like 027 and 176 may require further molecular analysis to guide outbreak prevention strategies (28). Given the increasing CDI outbreaks and insufficient surveillance in many regions, adopting a triple diagnostic algorithm—GDH test and ELISA with PCR confirmation for discordant results—would empower clinicians to achieve more accurate and timely CDI diagnoses, ultimately improving patient care and public health outcomes.