Introduction
Chronic Lymphoproliferative Disorders (CLPD) represent a diverse group of hematological malignancies characterized by the abnormal proliferation of lymphocytes. Accurate diagnosis and classification of CLPD are crucial for effective patient management, risk stratification, and tailored treatment strategies. Among the advanced diagnostic techniques, multiparameter flow cytometry has emerged as a cornerstone for differentiating between various CLPDs due to its high-throughput capabilities, rapid turnaround time, and logistical feasibility [1]. This technique allows for the simultaneous analysis of multiple cell surface markers, providing a comprehensive immunophenotypic profile essential for accurate Clpd Diagnosis and subtyping.
The differential diagnosis of CLPD encompasses entities like Chronic Lymphocytic Leukemia (CLL), hairy cell leukemia, and mantle cell lymphoma, each requiring distinct therapeutic approaches. Furthermore, within CLL itself, prognostic markers such as CD38 and ZAP70 expression are critical for assessing disease aggressiveness and predicting treatment response [2]. The increasing complexity of treatment modalities, including antibody therapies and stem cell transplantation, further underscores the need for precise and iterative evaluation of marker expression in CLPD.
Traditionally, CLPD diagnosis relies on a combination of absolute lymphocyte count, morphology, and immunophenotyping. Classical CLL diagnosis is defined by lymphocytosis (>5000/μL), the presence of small, mature lymphocytes with characteristic morphology [3], and a distinctive immunophenotype (typically CD5+, CD19+, CD23+, dim CD20, dim CD79b, dim sIg, and FMC7) [4]. However, distinguishing CLL from other CLPDs, particularly in resource-constrained settings, necessitates cost-effective diagnostic strategies. Optimizing antibody panels for flow cytometry to achieve accurate CLPD diagnosis while managing costs is a significant challenge.
Numerous studies have investigated the utility of various immunophenotypic markers and their combinations in CLPD diagnosis [5–7], aiming to refine diagnostic panels and minimize redundancy [8,9]. In economic settings where cost-effectiveness is paramount, such as in India, the development of minimal, yet informative, antibody panels is crucial. This study was undertaken at a tertiary cancer center in India to evaluate the effectiveness of a specific antibody panel for CLPD diagnosis, focusing on CLL, and to analyze the clinicopathological characteristics of CLPD patients in this region. This research aims to contribute to the optimization of cost-effective diagnostic approaches for CLPD, particularly in resource-limited settings, by highlighting the utility of a streamlined flow cytometry panel in achieving accurate and timely diagnoses.
Materials and Methods
Patients and Samples
This retrospective study included newly diagnosed and referred cases of CLPD at a multispecialty oncology hospital between November 2008 and December 2011. Diagnosis was based on integrated assessment of clinical data, morphology, immunophenotyping, and cytogenetics. Samples included EDTA-anticoagulated peripheral blood and bone marrow for morphology and immunophenotyping, and heparinized samples for cytogenetics. All samples were collected following standard aseptic procedures with informed consent obtained by the clinicians as part of routine diagnostic workup.
Morphological Analysis
Peripheral blood and bone marrow aspirate smears were stained with Leishman and May-Grunwald Giemsa stains, respectively. Morphological classification of cases into typical CLL, prolymphocytic leukemia (PLL), or other CLPD categories was based on established hematopathological guidelines [10] and absolute lymphocyte counts (>5×10^9/L for at least 3 months). Typical CLL morphology was defined by small lymphocytes with scant cytoplasm, condensed chromatin, and inconspicuous nucleoli, often accompanied by smudge cells. Cases with prolymphocyte counts between >10% and
Immunophenotyping by Flow Cytometry
Immunophenotyping was performed using a primary panel of monoclonal antibodies from Beckman Coulter, India. This panel included CD45, CD10, CD5, CD19, CD20, CD23, FMC7, and CD79b. Additional markers such as CD3, CD22, CD25, CD103, CD38, ZAP70, kappa, and lambda light chains were used for further characterization and differential diagnosis. Briefly, blood or bone marrow samples were incubated with antibodies for 30 minutes at room temperature, washed, and analyzed on a Dako CyAn ADP instrument. At least 10,000 events were acquired per analysis, and data were analyzed using SUMMIT® software (version 5.4). Antigen positivity was defined as ≥20% of cells expressing the marker. Expression levels were categorized as strong, moderate, or dim based on fluorescence intensity relative to isotype controls [11,12].
Cytogenetic Analysis
Cytogenetic studies were performed using reagents from Invitrogen, Bangalore, India. Short-term cultures (direct, 24h, and 48h) were initiated from fresh bone marrow aspirates in RPMI 1640 supplemented with L-Glutamine, fetal bovine serum, and penicillin-streptomycin. Cultures were treated with colcemid to arrest metaphase, followed by hypotonic treatment and fixation. G-banding was performed using trypsin and Giemsa stain. Karyotypes were described according to the International System for Human Cytogenetic Nomenclature. Fluorescence In Situ Hybridization (FISH) was performed using a CLL-specific diagnostic kit (MP Biomedical LLC, Mumbai, India) with probes for 13q14/p53 (17p13), 11q22/GLI 12q13, 6q21/8q24 C-myc, and IGH gene break apart. At least 200 cells were counted per probe set under a fluorescence microscope.
Results
Patient Demographics
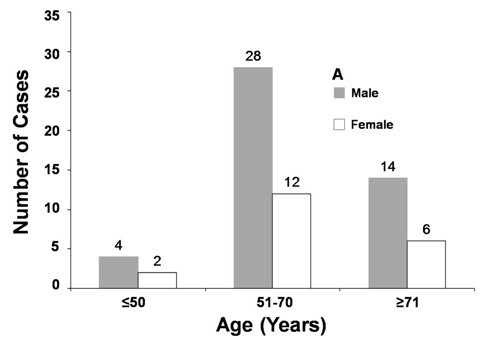
A total of 66 CLPD cases were analyzed, with a median age of 64.5 years (range 43-84 years). The majority of cases (60.6%) were in the 51-70 year age group. There was a significant male predominance, with a male to female ratio of 2.3:1 (69.6% males, 30.3% females). Specifically, in the cohort, 40 cases were diagnosed as CLL, and 9 as atypical CLL. The remaining 17 cases were classified as other CLPDs, including 13 Non-Hodgkin’s Lymphomas (NHL), 2 Hairy Cell Leukemias (HCL), 1 Follicular Lymphoma (FL), and 1 Prolymphocytic Leukemia (PLL).
Classification of CLPD Cases by Sex and Age
Morphological Findings
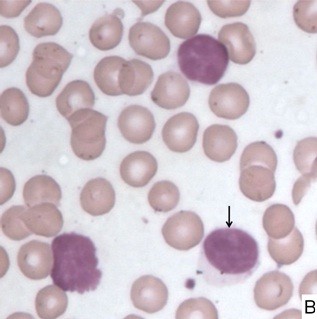
Morphological review, concurrent with flow cytometry immunophenotyping, was performed by two pathologists in all cases. Based on peripheral blood and bone marrow morphology and lymphocyte counts, cases were categorized as CLL, CLL/PLL, or other CLPD. Median leukocyte counts across age groups showed no significant association. Most cases were morphologically classified as CLL based on the predominance of small mature lymphocytes and the presence of smudge cells. Two cases were suspected HCL, and one PLL, based on morphology.
Median Leukocyte Counts of CLPD Cases by Age Group
Morphology of CLL Blood Smear (Low Power)
Morphology of CLL Lymphocyte (High Power)
Cytogenetic Abnormalities
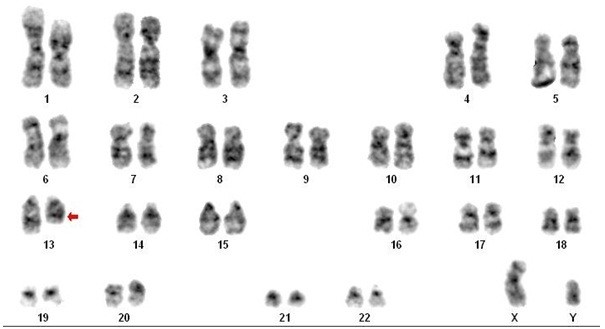
Cytogenetic analysis (karyotyping and FISH) was performed in 15 CLPD cases, revealing abnormalities in 3 (20%). One case showed t(14;18)(q32;q21), a hallmark of follicular lymphoma, consistent with morphology and CD5-/CD10+ immunophenotype. Another case demonstrated del(13), a common abnormality in CLL. The third case exhibited trisomy 12q, also frequently observed in CLL.
Karyotyping Results Showing del(13) in CLL
Immunophenotypic Profiles
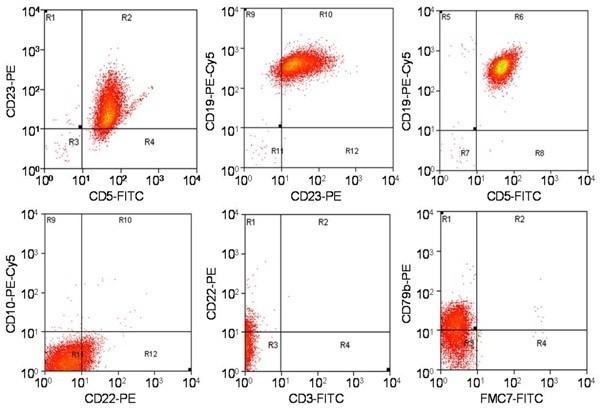
Immunophenotyping results, summarized in Table/Fig-5, showed that all 66 cases were CD19+ B-cells. In CLL cases, the majority (38/40) co-expressed CD5 and CD23. Two CLL cases were CD5 negative. CD3 and CD10 were positive in a small subset of CLL cases (10% and 5%, respectively), but morphology supported CLL diagnosis. Prognostic markers CD38 and ZAP70 were positive in 40% and 22.5% of CLL cases, respectively. Atypical CLL cases consistently expressed CD22, with two being CD5 negative. Among other CLPD cases, one follicular lymphoma was CD5-/CD10+. The PLL case was CD5-/CD23+/CD22+. Hairy cell leukemias (HCL) were CD5-/CD23-/CD22+/CD25+/CD103+. Remaining CLPD cases showed variable marker expression, requiring lymph node biopsy and immunohistochemistry for definitive diagnosis due to panel limitations.
Immunophenotypic Diagnostic Criteria for CLPD
| CD 19 | CD 5 | CD 23 | CD 20 | CD 22 | FMC 7 | CD 79b | CD 10 | CD 3 | CD 25 | CD 103 |
|---|---|---|---|---|---|---|---|---|---|---|
| CLL | + | + | + | + | – | -/(+) | -/(+) | – | – | – |
| MCL | + | + | – | + | + | + | + | – | – | – |
| HCL | + | – | – | + | + | + | + | – | – | + |
| FL | + | – | – | + | + | – | + | + | – | – |
| PLL | + | ± | ± | + | + | + | + | – | – | – |
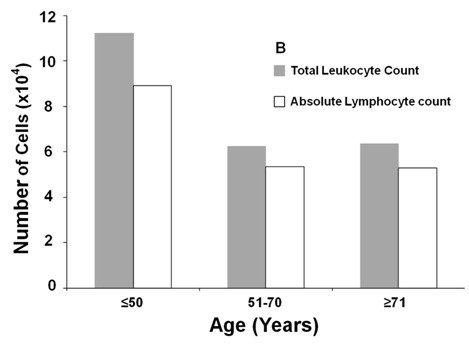
Immunophenotypic Profiles of CLPD Subtypes
| Antigen | CLL (Positive/Total) | Atypical CLL (Positive/Total) | CLPD (Positive/Total) |
|---|---|---|---|
| CD3 | 4/40 | 0/9 | 1/17 |
| CD5 | 38/40 | 2/9 | 6/17 |
| CD10 | 2/40 | 0/9 | 2/17 |
| CD19 | 40/40 | 9/9 | 17/17 |
| CD20 | 34/40 | 9/9 | 17/17 |
| CD22 | 0/40 | 9/9 | 16/17 |
| CD23 | 40/40 | 9/9 | 4/17 |
| CD25 | 0/40 | 0/9 | 2/17 |
| CD103 | 0/40 | 0/9 | 2/17 |
| CD38 | 16/40 | 1/9 | 11/17 |
| ZAP70 | 9/40 | 0/9 | 4/17 |
| CD79b | 23/40 | 3/9 | 11/17 |
| FMC7 | 5/40 | 2/9 | 9/17 |
Flow Cytometry Immunophenotype of CLL Case
CD20 was expressed in 85% of CLL cases, while CD79b and FMC7 were expressed in 57.5% and 12.5%, respectively. CD20 expression was consistent across atypical CLL and other CLPDs, while CD79b and FMC7 expression were higher in these groups (p=0.005). Lack of CD22 expression showed the highest correlation with CLL diagnosis (r2= -0.97, P=0.000), followed by CD5 (r2= 0.68, P=0.000) and CD23 (r2= 0.61, P=0.000) positivity. A significant negative correlation was found between CD23 and FMC7 expression (r2= -0.52, P=0.000).
Discussion
Accurate identification and classification of CLPD are essential for patient care. Diagnosis requires an integrated approach combining morphology, immunophenotyping, and cytogenetics. Flow cytometry immunophenotyping is particularly crucial for CLPD subtyping, especially when cytogenetic analysis is challenging or morphology alone is insufficient. Surface marker expression is also important for prognostication and monitoring disease progression post-therapy. Selecting cost-effective immunophenotypic panels that provide maximal diagnostic information is a key consideration for diagnostic laboratories.
This study evaluated a specific antibody panel for CLPD diagnosis, focusing on CLL, in an Indian tertiary cancer center. Epidemiological patterns of CLL occurrence were also investigated. CLL is the most common leukemia, predominantly affecting older adults, with a median age of diagnosis around 72 years in Western populations [19]. In this study, the median age was slightly lower at 64.5 years, with the highest incidence in the 51-70 age group, consistent with reports of a younger median age in Indian populations [20], possibly due to ethnic and genetic factors [21, 22]. The male predominance observed in this study aligns with global CLL epidemiology [24].
Morphological findings were consistent with typical CLL features. Immunophenotyping using the basic panel effectively diagnosed CLL in most cases, with the absence of CD22 expression being the most strongly correlated marker. CD5 and CD23 positivity also significantly supported CLL diagnosis. The panel also aided in identifying follicular lymphoma and hairy cell leukemia among other CLPDs. Higher expression of CD20, FMC7, and CD79b in non-CLL cases, particularly FMC7, suggests their utility in differentiating CLPD subtypes. The inverse correlation between CD23 and FMC7 expression warrants further investigation for clinical significance.
The findings suggest that a basic minimal panel, including CD45, CD20, CD22, CD79b, CD19, CD5, CD23, CD10, FMC7, CD3, CD5, CD25, CD103, kappa, and lambda, is sufficient for routine CLL diagnosis. However, comprehensive CLPD stratification requires expanded panels. A tiered approach, starting with a basic panel and expanding as needed, is recommended for cost-effective and accurate CLPD diagnosis.
Financial or Other Competing Interests
None declared.
References
[1] Muzzafar, T., and A. N. Jain. 2006. “Role of Flow Cytometry in Diagnosis of Hematological Malignancies.” Indian J Hematol Blood Transfus 22 (3): 74-79.
[2] Crespo, M., J. Bosch, X. Villamor, et al. 2003. “ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia.” N Engl J Med 348 (18): 1764-75.
[3] Bennett, J. M., D. Catovsky, M. T. Daniel, et al. 1989. “Proposals for the classification of chronic (mature) B and T and NK-cell leukaemias. French-American-British (FAB) Cooperative Group.” J Clin Pathol 42 (6): 567-84.
[4] Moreau, E. J., F. Matutes, A. A’Hern, et al. 1997. “Improvement of the WHO classification of lymphoid neoplasms: the immunophenotyping contribution.” Ann Oncol 8 Suppl 2: 49-54.
[5] ையானந்தನ್, K., and T. V. Sreeja. 2012. “Utility of CD23 and FMC-7 Markers in Diagnosing Chronic Lymphocytic Leukemia: An Analysis from a Tertiary Cancer Centre in South India.” J Clin Diagn Res 7 (7): 1366–1371.
[6] Rawstron, A. C., M. J. Davies, J. Das Gupta, et al. 2001. “Diagnostic and MRD applications of high-sensitivity flow cytometry in B-cell chronic lymphocytic leukaemia.” Leukemia 15 (4): 540-46.
[7] ையானந்தನ್, K., and T. V. Sreeja. 2011. “Diagnostic utility of CD23 and FMC-7 markers in B-cell chronic lymphocytic leukemia.” Indian J Pathol Microbiol 54 (1): 53-57.
[8] Van Dongen, J. J., E. R. van den Beemd, A. C. Rawstron, et al. 1999. “Minimal residual disease diagnostics in acute lymphoblastic leukemia: recommendations for test design and interpretation.” Leukemia 13 Suppl 2: 190-97.
[9] Wood, B. L., R. Arroz, P. Barnett, et al. 2006. “Minimal residual disease detection in chronic lymphocytic leukemia.” Cytometry B Clin Cytom 70 (2): 101-13.
[10] Swerdlow, S. H., E. Campo, N. L. Harris, et al. 2008. “WHO classification of tumours of haematopoietic and lymphoid tissues.” IARC Press.
[11] Gratama, J. W., A. C. Kraan, F. J. Bredius, et al. 1998. “Optimization of flow cytometric enumeration of CD34+ hematopoietic stem cells in peripheral blood and bone marrow.” Cytometry 34 (2): 59-71.
[12] Lanier, L. L., A. M. Le, C. I. Civin, et al. 1986. “Antigenic, functional, and molecular genetic studies of human natural killer cell clones.” J Immunol 136 (12): 4415-23.
[13] Tsujimoto, Y., J. Yunis, L. Onorato-Showe, et al. 1984. “Molecular cloning of the chromosomal breakpoint of B-cell lymphomas and leukemias with the t(14;18) chromosome translocation.” Science 224 (4656): 1403-6.
[14] Weinberg, D. S., and R. S. Neiman. 1987. “Mantle cell lymphoma. A neoplasm of B cells with intermediate differentiation features.” Am J Surg Pathol 11 (10): 782-90.
[15] Döhner, H., M. Stilgenbauer, P. Döhner, et al. 1997. “Genomic aberrations in chronic lymphocytic leukemia. II. Cytogenetic findings and their correlations with disease characteristics and prognosis.” Leukemia 11 (2): 192-99.
[16] Juliusson, G., G. Oscier, A. Fitchett, et al. 1990. “Prognostic subgroups in B-cell chronic lymphocytic leukemia defined by specific chromosomal abnormalities.” N Engl J Med 323 (11): 720-24.
[17] Foon, K. A., and R. F. Todd 3rd. 1986. “Immunologic classification of leukemia and lymphoma.” Blood 68 (1): 1-31.
[18] Ries, L. A. G., M. Eisner, C. Kosary, et al. 2006. SEER Cancer Statistics Review, 1975-2003. Bethesda, MD: National Cancer Institute.
[19] National Cancer Institute. 2010. “SEER Stat Fact Sheets: Leukemia.” Bethesda, MD: National Cancer Institute.
[20] Gogia, A., S. Raina, V. Sharma, et al. 2011. “Chronic lymphocytic leukemia in India: a retrospective analysis of clinical characteristics and treatment outcome.” Indian J Cancer 48 (1): 48-53.
[21] De Roos, A. J., M. E. Cerhan, P. Linet, et al. 2008. “Chronic lymphocytic leukemia and small lymphocytic lymphoma incidence patterns in North America and Europe.” Cancer Causes Control 19 (10): 1209-17.
[22] Goldman, R. H., and S. V. Smith. 2011. “Racial and ethnic disparities in chronic lymphocytic leukemia incidence and survival: a SEER database analysis.” Leuk Lymphoma 52 (11): 2077-85.
[23] Dhamne, C. A., S. Banavali, N. B. Parikh, et al. 2011. “Higher frequency of CD4+CD25high T regulatory cells in Indian patients with chronic lymphocytic leukemia is associated with poor prognosis.” Leuk Res 35 (8): 1024-29.
[24] SEER. 2023. “Cancer stat facts: leukemia.” National Cancer Institute. Accessed September 10, 2023. https://seer.cancer.gov/statfacts/html/leuks.html.