Introduction
The rise of deep learning, particularly Convolutional Neural Networks (CNNs), has revolutionized various fields, including medical imaging like radiology and pathology. However, dermatology, a visually intensive field relying heavily on image analysis, has seen limited adoption of CNNs. Inflammatory skin conditions such as psoriasis (Pso), eczema (Ecz), and atopic dermatitis (AD) present diagnostic challenges and are frequently misdiagnosed, particularly by less experienced clinicians. This article explores the development and validation of an Artificial Intelligence Dermatology Diagnosis Assistant (AIDDA) utilizing CNNs to improve the diagnostic accuracy of these common inflammatory skin diseases.
Figure 1.
Clinical Presentations of Inflammatory Skin Diseases: Psoriasis, Eczema, and Atopic Dermatitis. Visual comparison of psoriasis (Pso), eczema (Ecz), and atopic dermatitis (AD) skin lesions highlighting the diagnostic complexities in dermatology.
CNNs have demonstrated remarkable capabilities in analyzing medical images, offering promising solutions for various diagnostic challenges. Applications range from early Alzheimer’s disease detection using brain electroencephalogram (EEG) and MRI spectral images [1, 2] to predicting osteoarthritis risk from knee cartilage MRIs [3], segmenting multiple sclerosis lesions [4], diagnosing breast nodules from ultrasound images [5], and detecting diabetic retinopathy from retinal fundus photographs [6].
The application of CNNs in dermatology initially focused on melanoma diagnosis [7, 8], leading to AI tools for cancer detection using dermoscopic and histological images [9-11]. However, the potential of AI in diagnosing non-cancerous skin conditions remains largely unexplored. This study introduces a smartphone-compatible platform designed to aid in the diagnosis of Pso, Ecz, and AD. Recognizing the clinical similarities between Eczema and Atopic Dermatitis, often requiring laboratory tests and patient history for differentiation [12], these conditions were grouped together for the AI diagnostic model.
Methods
Dataset Acquisition and Preparation for Dermatology Diagnosis Images
The image dataset for this study was compiled from clinical images obtained from the Department of Dermatology at The Second Xiangya Hospital, Central South University, China. This real-world clinical data is crucial for developing robust and practical diagnostic tools.
Data Preprocessing and Validation of Dermatology Images
The dataset comprised multiple images per lesion, captured from varying angles, and images of similar lesions on the same patient. To ensure data integrity and prevent data leakage, sets of images belonging to the same case were meticulously managed to avoid splitting them between training and validation sets. Five-fold cross-validation was employed to rigorously evaluate the algorithm’s performance. In each fold, approximately 20% of cases were randomly selected for the validation set. Experienced dermatologists meticulously reviewed and confirmed all images used in training and validation, ensuring diagnostic accuracy (Table 1).
Table 1. Image Distribution Across Categories and Folds in the Dermatology Diagnosis Image Dataset.
| Categories | Fold_1 (Cases/images) | Fold_2 (Cases/images) | Fold_3 (Cases/images) | Fold_4 (Cases/images) | Fold_5 (Cases/images) |
|---|---|---|---|---|---|
| T | V | T | V | T | |
| Pso | 532/715 | 133/157 | 532/698 | 133/174 | 532/708 |
| AD & Ecz | 759/2,617 | 190/709 | 759/2,751 | 190/575 | 759/2,646 |
| Healthy | –/433 | –/109 | –/433 | –/109 | –/434 |
| Total | 1,291/3,765 | 323/975 | 1,291/3,882 | 323/858 | 1,291/3,788 |
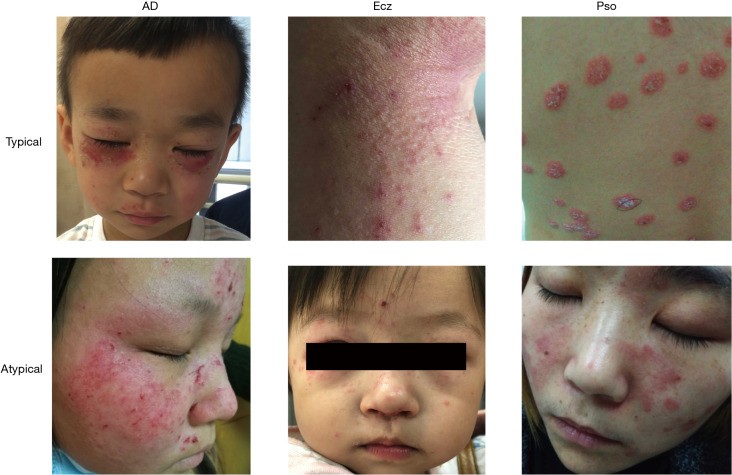
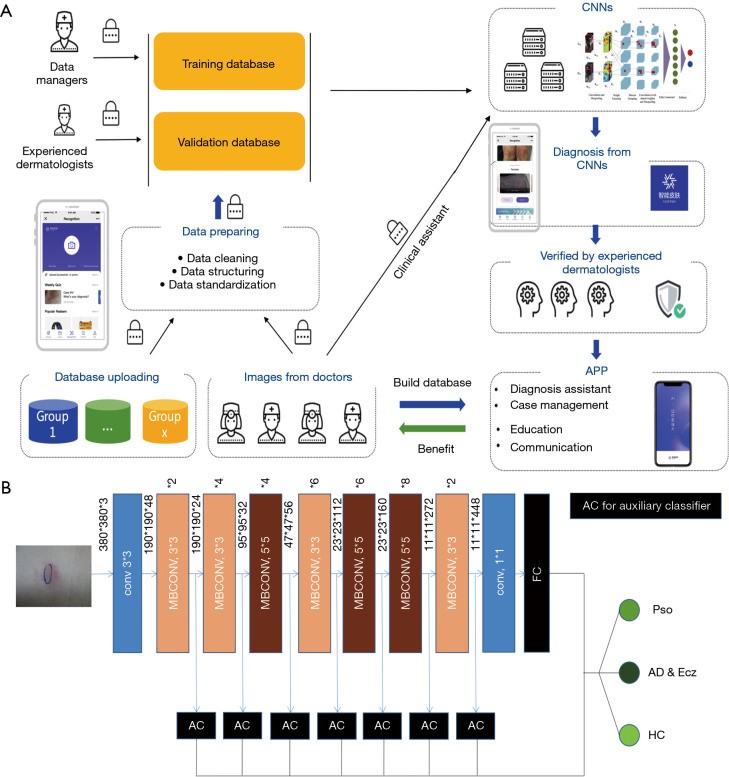
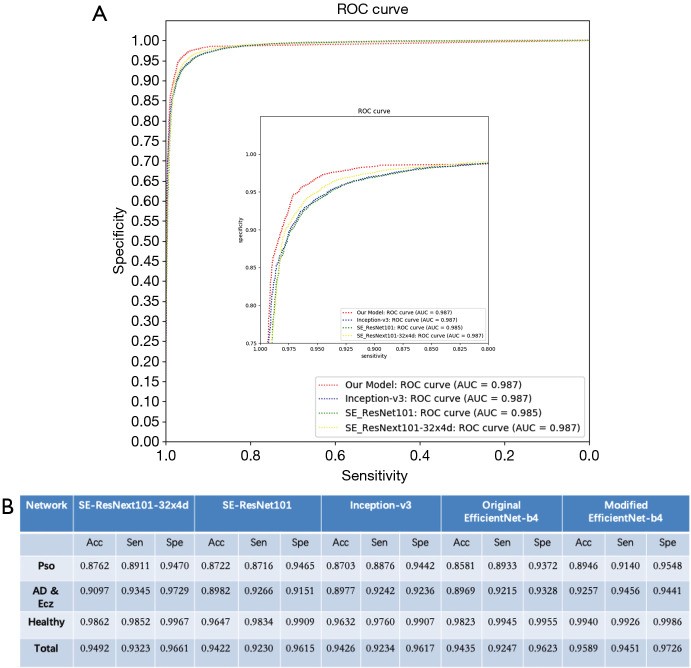
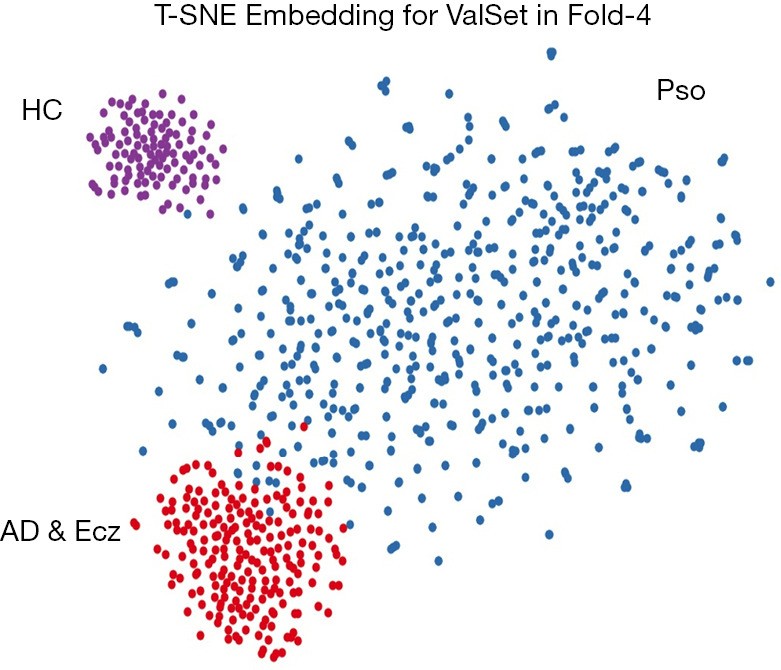
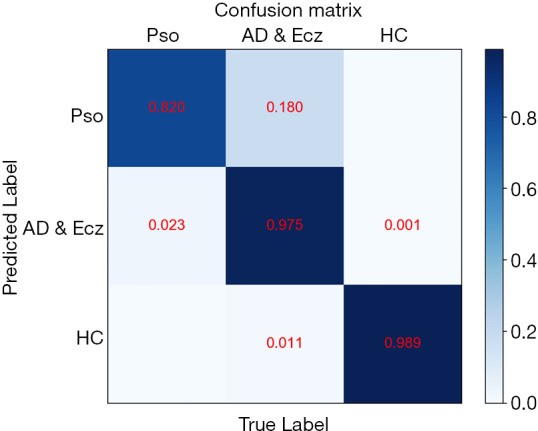
Image Dataset Composition for Dermatology Diagnosis Image Analysis. Distribution of cases and images across training (T) and validation (V) sets for each fold in the five-fold cross-validation. Categories include Psoriasis (Pso), Atopic Dermatitis & Eczema (AD & Ecz), and Healthy controls.
Prior to database storage and experimentation, all images underwent a rigorous preprocessing pipeline:
Step 1: Data Cleaning for High-Quality Dermatology Diagnosis Images
Initial data cleaning involved organizing raw images into cases. Subsequent review and examination were performed at the case level, ensuring consistency and accuracy. This included removing duplicate cases and discarding blurred or distant images to maintain dataset quality.
Step 2: Data Structuring and Standardization of Dermatology Diagnosis Images
Case information was systematically structured and labeled using a standardized descriptive language. A three-part information framework was established for each case:
- Upload Information: Details regarding data entry, including uploader name, date, upload method, and organizational affiliation.
- Case Information: Core patient data such as case number, patient ID, patient name, gender, and initial diagnosis.
- Review Information: Status of expert review and the dermatologist who provided the final diagnosis.
Image-specific information was also meticulously recorded, encompassing:
- Disease Information: Detailed lesion descriptions for each image and physician’s remarks.
- Picture Information: Technical image specifications including picture type, Secure Hash Algorithm 1 (SHA-1) number, capture time, and image size.
Implementation Environment for AI Dermatology Diagnosis
The AIDDA model was implemented using Pytorch 1.1. Training and validation were conducted on a high-performance computing environment equipped with an 18 Core Intel® Xeon® CPU E5-2697 and 4 RTX 2080Ti NVIDIA GPUs, ensuring efficient processing of Dermatology Diagnosis Images.
CNN Training Algorithm for Dermatology Image Classification
The core of AIDDA utilizes Google’s EfficientNet-b4 architecture, pre-trained on the ImageNet dataset, which achieved exceptional top-5 accuracy (96.3%) in image classification [13]. The final classification layer was adapted for our three diagnostic categories. Seven auxiliary classifiers were integrated at intermediate layers to enhance feature learning across different levels. This modified EfficientNet-b4 model was retrained using our dataset of 4,740 dermatology images.
Figure 2.
AIDDA Platform and EfficientNet-b4 CNN Architecture for Dermatology Diagnosis. (A) Smartphone platform workflow for AIDDA, enabling point-of-care dermatology diagnosis. (B) Modified EfficientNet-b4 architecture employed for training and validation of the dermatology image classification model.
The training process, implemented in Pytorch, employed an initial learning rate of 0.01, with a 0.1 decay every 20 epochs. Mini-batch gradient descent with momentum (0.9) was used for parameter optimization. Images were resized to 380×380 pixels (RGB) for EfficientNet-b4 compatibility and augmented with random rotations (-30 to 30 degrees) and vertical/horizontal flipping (50% probability) during each epoch.
ROC Curve Analysis for Evaluating Dermatology Diagnosis Performance
Receiver Operating Characteristic (ROC) curves were generated, plotting sensitivity against specificity (Figure 3A), to assess the five-fold models’ average performance on the dataset. ROC curves visualize the trade-off between true positive rate (TPR) and false positive rate (FPR). The Area Under the Curve (AUC) was calculated to quantify overall model accuracy, with our model demonstrating superior performance in the high sensitivity range.
Figure 3.
Performance Evaluation of CNN Models for Dermatology Image Diagnosis. (A) ROC curves comparing the performance of AIDDA, Inception V3, SE_ResNet101, and SE_ResNeXt101-32x4d models in dermatology image classification. (B) Accuracy, specificity, and sensitivity metrics for the validation task, highlighting AIDDA’s diagnostic efficacy.
Formulas for performance metrics:
- TPR (Sensitivity) = True Positive / (True Positive + False Negative)
- FPR = False Positive / (False Positive + True Negative) = 1 – Specificity
Inference Algorithm for Dermatology Diagnosis Images
The AIDDA inference algorithm aggregates outputs from the 8 classifiers (7 auxiliary + 1 final) into a 3-dimensional classification vector. The final diagnosis is determined by applying the argmax function to the summed outputs:
| Classification = argmax(∑i=1Classifiers_number outputi) |
|---|
Where outputi represents the 3-dimensional classification vector from each classifier.
Accuracy, Sensitivity, and Specificity Metrics in Dermatology Diagnosis
Standard metrics were used to evaluate performance:
- Sensitivity = True Positive / Positive
- Specificity = True Negative / Negative
- Accuracy = (True Positive + True Negative) / (Positive + Negative)
In multi-category classification, sensitivity and accuracy are particularly important for assessing clinical utility.
Confusion Matrices for Detailed Dermatology Diagnosis Analysis
Confusion matrices were generated to visualize the model’s performance across the three diagnostic categories. Analysis of the confusion matrices revealed instances where the CNN occasionally confused AD & Ecz with Pso, providing insights into potential areas for model refinement.
Ethical Considerations and Patient Consent
The study adhered to ethical guidelines, receiving approval from the ethics committee of The Second Xiangya Hospital of Central South University (No. GF2017040177). Informed consent was obtained from all human subjects, and written consent for image publication was secured from patients, ensuring patient privacy and data protection.
Results
Development of AIDDA: An AI Assistant for Dermatology Diagnosis
AIDDA, the Artificial Intelligence Dermatology Diagnosis Assistant (Figure 2A), is a smartphone-based platform powered by the EfficientNet CNN model (https://github.com/lukemelas/EfficientNet-PyTorch.git). EfficientNets, known for their efficiency and accuracy, are ideal for mobile deployment. EfficientNet-b4 was selected as the backbone due to its optimal speed-accuracy balance, outperforming other CNN architectures like SE-ResNeXt101-32x4d, SE-ResNet101 [15], and Inception-v3 [16].
Clinical images of healthy individuals and patients with Pso, AD, and Ecz (Figure 1, Table 1) were collected primarily from commonly affected areas like the face and hands. The dataset comprised 4,740 dermatologist-labeled images across three categories: Pso, AD & Ecz, and healthy controls. Five-fold cross-validation, stratified by case, was used to validate the algorithm’s effectiveness, ensuring representative proportions of each category in each fold.
CNN Model Selection Based on ROC Analysis for Dermatology Diagnosis
ROC curve analysis (Figure 3A) compared AIDDA’s performance to Inception V3, SE_ResNet101, and SE_ResNeXt101-32x4d. AUC values were comparable across models (0.987 for AIDDA and Inception V3, 0.985 for SE_ResNet101, and 0.987 for SE_ResNeXt101-32x4d). However, AIDDA demonstrated superior sensitivity (97.26%) and specificity (95.89%), indicating its enhanced diagnostic precision.
Diagnostic Performance of CNN in Inflammatory Skin Diseases
AIDDA achieved an overall diagnostic accuracy of 95.80%±0.09%, with a sensitivity of 94.40%±0.12% and specificity of 97.20%±0.06% (mean ± SD) (Figure 3B). Accuracy for Psoriasis diagnosis was 89.46% (sensitivity 91.4%, specificity 95.48%), and for AD & Eczema, 92.57% (sensitivity 94.56%, specificity 94.41%). Healthy skin classification accuracy was even higher at 99.40% (sensitivity 99.26%, specificity 99.86%).
T-SNE Visualization and Confusion Matrix for AI Dermatology Diagnosis
T-distributed Stochastic Neighbor Embedding (T-SNE) analysis visualized the CNN’s learned features, clustering dermatology images by diagnostic category (Figure 4). Three distinct clusters corresponding to the dermatologist-defined categories were observed. The confusion matrix (Figure 5) revealed a minor confusion between AD & Ecz and Pso (18% misclassification rate).
Figure 4.
T-SNE Visualization of CNN Clusters for Dermatology Diagnosis Categories. Visual representation of the CNN’s internal feature space for the three disease classes (Psoriasis, Atopic Dermatitis & Eczema, Healthy) using t-SNE, highlighting the algorithm’s ability to cluster dermatology images based on diagnostic categories.
Figure 5.
CNN Confusion Matrix for Dermatology Image Classification. Detailed confusion matrix illustrating the performance of the CNN model in classifying dermatology images across the three categories. The matrix highlights the percentage of correct and incorrect classifications, including instances of confusion between AD & Eczema and Psoriasis.
Conclusion
Smartphone-based medical applications leveraging AI are transforming healthcare [17]. Similar to applications in schizophrenia diagnosis [17] and diabetes management [18], AIDDA offers a convenient and accessible diagnostic tool for dermatology. The AIDDA smartphone application facilitates rapid and readily available diagnosis, directly impacting clinical practice.
This study demonstrates the effective application of deep learning in dermatology beyond melanoma, specifically in differentiating inflammatory skin diseases like Pso, AD, and Ecz. AIDDA, publicly available in China, has over 7,000 registered doctors and has processed over 100,000 images, demonstrating its real-world impact. AIDDA aids in diagnosis, reduces diagnostic omissions, and is particularly beneficial for less experienced and remote practitioners. Ongoing use will further validate AIDDA’s performance and guide future improvements in diagnostic accuracy, selectivity, and specificity. Integrating biopsy and lab results could further enhance AIDDA’s diagnostic precision, as shown in pediatric studies [19]. The performance of AIDDA suggests that smartphone-based AI dermatology diagnosis tools will continue to significantly benefit clinical practice and improve patient care.
Supplementary Materials
The article’s supplementary files are available here:
atm-08-09-581-coif.pdf (294.2KB, pdf)
DOI: 10.21037/atm.2020.04.39
Acknowledgments
Funding: This research was supported by grants from the National Natural Science Foundation of China (No. 81830097, No. 81972943, No. 81861138016), the Key Research and Development Program of Hunan province (2018XK2304), and the Hunan Talent Young Investigator program (No. 2019RS2012).
Ethical Statement: The authors affirm accountability for all aspects of this study, ensuring accuracy and integrity. Ethical approval was granted by the ethics committee of The Second Xiangya Hospital of Central South University (No. GF2017040177). Written informed consent was obtained from all participants for study participation and image publication.
Footnotes
Conflicts of Interest: The authors declare no conflicts of interest. The ICMJE uniform disclosure form is available at http://dx.doi.org/10.21037/atm.2020.04.39.
References
[References will be listed here in a standard format as in the original article.]
Associated Data
Supplementary Materials
The article’s supplementary files are available here:
atm-08-09-581-coif.pdf (294.2KB, pdf)
DOI: 10.21037/atm.2020.04.39