1. Introduction to Dengue Diagnosis
The accurate and timely Laboratory Diagnosis of dengue virus infection is paramount. It plays a critical role in various aspects of healthcare and public health, including informing clinical care decisions, especially for early identification of severe dengue cases. Laboratory confirmation is essential for differentiating dengue from other febrile illnesses with similar symptoms, which is crucial for effective patient management. Beyond individual patient care, laboratory diagnosis underpins robust disease surveillance, effective outbreak control strategies, in-depth pathogenesis studies, advancements in academic research, vaccine development, and the rigorous conduct of clinical trials.
Diagnostic approaches for dengue infection in the laboratory encompass a range of methods, each designed to detect different components of the infection. These methods primarily focus on identifying the virus itself, its genetic material (viral nucleic acid), viral antigens, or the host’s immunological response in the form of antibodies. Often, a combination of these techniques is employed to provide a comprehensive and accurate diagnosis.
In the initial phase of dengue illness, typically within the first 4 to 5 days after the onset of symptoms, the dengue virus is actively circulating and can be detected in various bodily fluids and tissues. Serum, plasma, circulating blood cells, and other tissue samples are all potential sources for virus detection during this acute phase. Diagnostic strategies during this early stage often involve virus isolation, nucleic acid detection, or antigen detection to confirm the infection.
As the acute phase of the infection subsides, and the body’s immune system begins to mount a response, serological methods become the preferred diagnostic approach. Serology focuses on detecting and measuring antibodies produced by the body in response to the dengue virus. These antibodies, particularly IgM and IgG, provide valuable information about the stage of infection and the patient’s immune history.
Understanding the antibody response in dengue infection is further complicated by the host’s prior exposure to flaviviruses. Individuals experiencing a primary dengue infection (their first encounter with any flavivirus, or without prior flavivirus vaccination) exhibit a characteristic primary antibody response. This response is marked by a gradual increase in specific antibodies, starting with IgM.
Alt text: Timeline illustrating primary and secondary dengue infections and suitable diagnostic methods, highlighting IgM and IgG antibody responses and viral detection windows.
IgM antibodies are typically the first to appear in detectable levels. They are often present in about 50% of patients between days 3 to 5 of illness onset, increasing to approximately 80% by day 5 and reaching 99% by day 10. IgM levels peak around two weeks after symptom onset and then gradually decline, usually becoming undetectable within 2 to 3 months. IgG antibodies, on the other hand, appear later in primary infections. Anti-dengue serum IgG is generally detectable at low levels towards the end of the first week of illness, increasing slowly thereafter. Importantly, IgG antibodies can persist for many months, and possibly even for life, providing long-term immunological memory.
In contrast to primary infections, secondary dengue infections (in individuals previously infected with dengue or another flavivirus, or vaccinated against flaviviruses) elicit a different antibody response. Secondary infections are characterized by a rapid surge in antibody titers. These antibodies often exhibit broader reactivity across multiple flaviviruses. IgG is the dominant antibody isotype in secondary infections, reaching high levels even in the acute phase and persisting for extended periods, from 10 months to life. Interestingly, IgM levels in the early convalescent stage of secondary infections are typically lower compared to primary infections and might even be undetectable depending on the diagnostic test used. To differentiate between primary and secondary dengue infections, laboratories often employ IgM/IgG antibody ratios, which have become more prevalent than the traditional haemagglutination-inhibition test (HI).
The array of available laboratory diagnostic methods for dengue is extensive, designed to cater to diverse needs in patient care and disease control. The selection of a specific diagnostic method is influenced by several factors. These include the intended purpose of testing, such as clinical diagnosis, epidemiological surveillance, or vaccine development. The availability of laboratory facilities, the level of technical expertise, cost considerations, and the timing of sample collection relative to the illness stage also play crucial roles in method selection.
Generally, diagnostic tests that offer high sensitivity and specificity often require more sophisticated technologies and specialized technical skills. Conversely, rapid diagnostic tests, while offering ease of use and speed, may compromise on sensitivity and specificity. Techniques like virus isolation and nucleic acid detection, while being more labor-intensive and costly, generally offer higher specificity compared to antibody detection methods based on serology.
Alt text: Graph comparing dengue diagnostic tests based on accessibility and confidence, showing trade-offs between ease of use and result reliability.
An important consideration is the inverse relationship between the ease of use or accessibility of a dengue diagnostic method and the confidence level in the test results. More accessible and rapid tests might sacrifice some degree of accuracy compared to more complex and laboratory-based methods. Understanding these trade-offs is crucial for selecting the most appropriate diagnostic strategy in different clinical and public health scenarios.
2. Key Considerations in Choosing Dengue Diagnostic Methods
2.1. Relevance to Clinical Management
Dengue virus infection presents with a wide spectrum of clinical manifestations, many of which are non-specific and overlap with other febrile illnesses. This clinical ambiguity makes diagnosis based solely on clinical symptoms unreliable. Therefore, early laboratory confirmation of a clinical suspicion of dengue is highly valuable. This is particularly important because dengue can progress rapidly in some individuals from mild symptoms to severe disease, and potentially to death. Early and accurate diagnosis allows for timely clinical intervention, which can be life-saving, especially in severe cases.
In the early stages of illness, specifically before day 5 during the febrile period, several laboratory methods are suitable for diagnosing dengue infections. These include virus isolation in cell culture, detection of viral RNA using nucleic acid amplification tests (NAAT), and detection of viral antigens using enzyme-linked immunosorbent assays (ELISA) or rapid diagnostic tests.
Virus isolation in cell culture is a highly specific method but is typically restricted to laboratories with specialized infrastructure and experienced personnel. For successful virus culture, maintaining the integrity of blood samples is critical. Samples must be kept cooled or frozen during transport from the patient to the laboratory to preserve virus viability. The process of virus isolation and subsequent identification in cell cultures usually takes several days to complete.
Nucleic acid detection assays, particularly those with high performance characteristics, can detect dengue viral RNA within a shorter timeframe of 24 to 48 hours. However, these tests necessitate expensive equipment and reagents and require strict quality control procedures to prevent contamination. They must be performed by highly trained and experienced technicians to ensure accuracy and reliability.
NS1 antigen detection kits have become commercially available and offer a more accessible option for laboratories with limited resources. These kits can provide results within a few hours. Rapid dengue antigen detection tests are particularly advantageous in field settings and can yield results in less than an hour. Currently, most of these assays are not serotype-specific and are undergoing evaluations for their diagnostic accuracy and cost-effectiveness in various clinical and epidemiological contexts.
Alt text: Table summarizing dengue diagnostic methods, outlining operating characteristics such as sensitivity, specificity, turnaround time, and comparative costs.
After day 5 of illness, the diagnostic landscape shifts as dengue viruses and antigens typically disappear from the bloodstream, coinciding with the rise of specific antibodies. While NS1 antigen may persist for a few days after the fever subsides in some patients, serological tests become increasingly relevant for diagnosis from this point onwards. Dengue serologic tests are generally more widely available in dengue-endemic countries compared to virological tests. Furthermore, specimen transport for serology is less stringent as immunoglobulins are stable at tropical room temperatures, simplifying logistics.
For serological diagnosis, the timing of specimen collection is more flexible compared to virus isolation or RNA detection. Antibody responses can be effectively measured by comparing samples collected during the acute phase of illness with convalescent samples taken weeks or months later. However, it’s important to note that in some secondary dengue infections, the detectable IgM response may be low or absent, potentially reducing the diagnostic accuracy of IgM ELISA tests.
Rapid diagnostic tests for serology can provide results in under an hour, offering quick turnaround. However, caution is advised when relying solely on rapid tests for dengue diagnosis. The performance of all commercially available rapid tests has not yet been fully evaluated by reference laboratories, and their sensitivity and specificity can vary.
A significant serological finding indicative of acute or recent flavivirus infection is a four-fold or greater increase in antibody levels measured by IgG ELISA or the haemagglutination inhibition (HI) test in paired sera (acute and convalescent). However, waiting for convalescent serum collection, typically at patient discharge, is often not clinically practical for immediate diagnosis and management. It provides a retrospective diagnosis rather than guiding acute clinical decisions.
2.1.1. Differential Diagnosis Considerations
Dengue fever can be easily mistaken for other illnesses, particularly in non-epidemic settings. This diagnostic challenge is further complicated by the geographical origin of the patient, as various regions have different prevalent diseases that can mimic dengue. When considering differential diagnosis, it is crucial to rule out other etiologies, including infections caused by other flaviviruses. These include yellow fever, Japanese encephalitis, St. Louis encephalitis, Zika virus, and West Nile virus. Additionally, alphavirus infections, such as those caused by Sindbis virus and chikungunya virus, should be considered.
Beyond viral infections, other causes of fever must be excluded in the differential diagnosis of dengue. These include malaria, leptospirosis, typhoid fever, and Rickettsial diseases (such as infections caused by Rickettsia prowazeki, R. mooseri, R. conori, R. rickettsii, Orientia tsutsugamushi, Coxiella burnetii, etc.). Other infectious diseases like measles, enterovirus infections, influenza, and influenza-like illnesses also share symptomatic overlap with dengue and need to be considered. In regions where they are prevalent, haemorrhagic fevers caused by Arenaviridae (e.g., Junin virus), Filoviridae (e.g., Marburg and Ebola viruses), and Bunyaviridae (e.g., hantaviruses, Crimean-Congo haemorrhagic fever virus) must also be considered in the differential diagnosis, particularly in severe cases.
For accurate dengue diagnosis, a combined approach of identifying the virus, viral RNA, or viral antigen, alongside detecting the antibody response, is preferable to relying on either approach alone. This integrated diagnostic strategy enhances the sensitivity and specificity of dengue confirmation.
Alt text: Table outlining the interpretation of dengue diagnostic tests, adapted from the DENCO study, showing how different test results correlate with infection status.
Despite the advancements in dengue diagnostics, an ideal diagnostic test that fulfills all desirable criteria—early and rapid diagnosis, affordability for diverse healthcare systems, ease of use, and robust performance—is still not available. Ongoing research and development efforts are crucial to bridge this gap and improve dengue diagnostic capabilities worldwide.
2.2. Application in Outbreak Investigations
During dengue outbreaks, the clinical presentation of patients can vary significantly depending on the stage of the outbreak and the individual’s disease progression. Some patients may present with fever, with or without rash, during the acute illness phase. Others might exhibit signs of plasma leakage or shock, indicative of severe dengue. Haemorrhagic manifestations may be apparent in some cases, while others might only be identified during the convalescent phase.
In a suspected dengue outbreak, one of the foremost priorities is to rapidly identify the causative agent. This is critical for implementing appropriate public health measures to control the outbreak and for guiding physicians in initiating appropriate clinical management for acutely ill patients. In outbreak scenarios, the speed and specificity of diagnostic tests are generally more critical than test sensitivity. Rapid and accurate diagnosis is essential for informing immediate public health responses and clinical strategies.
Samples collected from febrile patients during an outbreak can be tested using nucleic acid amplification methods in well-equipped laboratories. Alternatively, a broader range of laboratories can utilize ELISA-based dengue antigen detection kits for quicker results. If specimens are collected after day 5 of illness, commercial IgM ELISA or sensitive dengue IgM rapid tests can provide suggestive evidence of a dengue outbreak. However, it is preferable to confirm these initial findings with more reliable serological tests performed in a reference laboratory with comprehensive arbovirus diagnostic capabilities. Serological assays are particularly useful in determining the geographical extent and magnitude of outbreaks, aiding in epidemiological mapping and resource allocation.
2.3. Role in Surveillance Systems
Dengue surveillance systems are designed to monitor the circulation of dengue viruses within both human and mosquito populations. Effective surveillance is crucial for early detection of outbreaks, monitoring disease trends, and evaluating the impact of control measures. The diagnostic tools employed in dengue surveillance must be highly sensitive and specific to accurately detect and identify dengue viruses, and they must also be affordable for the country’s healthcare budget to ensure sustainable and widespread surveillance efforts.
Laboratories responsible for dengue surveillance are typically national and/or regional reference laboratories. These specialized laboratories possess the capacity to perform a wide range of diagnostic tests, including those described for clinical diagnosis and outbreak investigation. Furthermore, they often have the expertise and resources to diagnose a broad spectrum of other etiologies that may cause similar febrile illnesses, ensuring comprehensive surveillance and differential diagnosis.
2.4. Utility in Vaccine Trials
Vaccine trials are essential for assessing the safety and efficacy of dengue vaccines. Laboratory diagnostic methods play a crucial role in these trials, primarily to measure vaccine-induced immune responses and to confirm dengue cases in trial participants. The plaque reduction neutralization test (PRNT) and microneutralization assays are commonly used to measure neutralizing antibodies, which are key correlates of protection against dengue infection.
Following primary dengue infections in individuals without prior flavivirus immunity, neutralizing antibodies measured by PRNT tend to be relatively specific to the infecting dengue virus serotype. PRNT is considered the gold standard assay for measuring the titer of neutralizing antibodies in serum. It directly assesses the functional ability of antibodies to neutralize virus infectivity, thus providing a reliable measure of protection against infection. The principle of PRNT is based on the inactivation of the virus by neutralizing antibodies, preventing it from infecting and replicating in target cells.
However, after a secondary dengue virus infection, the antibody response becomes more complex. High-titer neutralizing antibodies are produced that can cross-react against multiple dengue serotypes, and often against non-dengue flaviviruses as well. This cross-reactivity arises from memory B-cells that produce antibodies targeting viral epitopes shared among different flaviviruses. Interestingly, during the early convalescent stage after sequential dengue infections, the highest neutralizing antibody titer is often directed against the first infecting virus serotype, rather than the most recent one. This phenomenon is known as “original antigenic sin” or antibody-dependent enhancement, which has implications for vaccine design and interpretation of serological responses in vaccine trials.
Despite its reliability, PRNT has drawbacks. It is labor-intensive and time-consuming, limiting its scalability for large vaccine trials. To address this, some laboratories have developed high-throughput neutralization tests for use in large-scale surveillance studies and vaccine trials. However, variability in PRNT results across different laboratories has been observed. Standardization of PRNT protocols is crucial to minimize inter-laboratory variations. This includes using standardized cell lines, consistent virus strains, and controlled incubation temperatures and times for virus-antibody interactions. Careful calculation of input virus is also important to avoid plaque overlap and ensure accurate quantification. Mammalian cell lines, such as VERO cells, are generally recommended for producing seed viruses used in PRNT.
The microneutralization assay operates on a similar principle to PRNT but is designed to be more efficient and high-throughput. Various modifications of microneutralization assays exist. One variation involves staining viral antigen using a labeled antibody and measuring the quantity of antigen colorimetrically, instead of counting plaques. Nucleic acid measurement using PCR can also be incorporated into microneutralization assays. Microneutralization assays are designed to use smaller reagent volumes and to facilitate testing larger numbers of samples, making them more suitable for large-scale vaccine trials and seroepidemiological studies. In viral antigen detection-based microneutralization tests, the incubation time after infection must be standardized to avoid measuring growth after multiple replication cycles, as the spread of virus is not limited as in PRNTs using semisolid overlays. Incubation periods need to be virus-specific because different viruses grow at different rates. Similar to standard PRNTs, antibodies measured by microneutralization assays in individuals with secondary infections may exhibit broad cross-reactivity with all four dengue serotypes.
In drug trials for dengue therapeutics, confirmed etiological diagnosis is essential for patient enrollment and outcome assessment. Diagnostic criteria for “highly suggestive” and “confirmed” dengue cases are crucial in these trials to ensure the study population is accurately defined and treatment efficacy can be reliably evaluated.
Alt text: Table summarizing the advantages and limitations of various dengue diagnostic methods, considering factors like turnaround time, cost, expertise required, and sensitivity.
Each dengue diagnostic method has its own set of advantages and limitations, influencing its suitability for different purposes. These factors must be carefully considered when selecting diagnostic strategies for clinical management, outbreak control, surveillance, and vaccine and drug development.
3. Current Dengue Diagnostic Methodologies
3.1. Virus Isolation Techniques
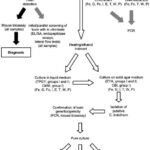
Virus isolation is a fundamental technique in virology, and for dengue, it involves culturing the virus from patient samples to confirm infection and for research purposes. Specimens for virus isolation should ideally be collected early in the course of infection, during the period of viremia, which is typically before day 5 of illness onset. During this time, the virus is most likely to be present in detectable quantities in the patient’s body.
The primary sources for virus isolation are serum, plasma, and peripheral blood mononuclear cells (PBMCs). In fatal cases, attempts can also be made to isolate the virus from tissues collected at autopsy, such as liver, lung, lymph nodes, thymus, and bone marrow. These tissues can provide valuable insights into viral tropism and pathogenesis.
Dengue virus is known to be heat-labile, meaning it is sensitive to heat and can degrade if not properly handled. Therefore, specimens intended for virus isolation must be handled with care to maintain virus viability. While awaiting transport to the laboratory, specimens should be refrigerated or packed in wet ice to keep them cold. For short-term storage up to 24 hours, maintaining a temperature between +4 °C and +8 °C is recommended. For longer storage, specimens should be frozen at -70 °C in a deep freezer or stored in liquid nitrogen for optimal preservation. Storing specimens at -20 °C, even for short periods, is not recommended as it can compromise virus viability.
Cell culture is the most widely used method for dengue virus isolation in laboratories. The mosquito cell line C6/36, cloned from Aedes albopictus, or AP61, a cell line from Aedes pseudoscutellaris, are the preferred host cells for routine dengue virus isolation. These mosquito cell lines are highly susceptible to dengue virus infection and support efficient viral replication. However, it’s important to note that not all wild-type dengue viruses induce a cytopathic effect (CPE) in mosquito cell lines. CPE refers to visible changes in cell morphology due to viral infection. Therefore, even if CPE is not observed, cell cultures must be screened for specific evidence of dengue infection.
Screening is typically done using an antigen detection immunofluorescence assay. This assay utilizes serotype-specific monoclonal antibodies and flavivirus group-reactive or dengue complex-reactive monoclonal antibodies to detect dengue viral antigens within the infected cells. Immunofluorescence allows for the visualization of viral antigens under a microscope, confirming virus presence and identity.
While mosquito cell lines are preferred, several mammalian cell cultures, such as Vero, LLCMK2, and BHK21 cells, can also be used for dengue virus isolation. However, they are generally less efficient than mosquito cell lines for primary isolation. Virus isolation followed by immunofluorescence assay confirmation typically requires 1 to 2 weeks to complete. The success of virus isolation is highly dependent on proper specimen transport and storage to preserve virus viability.
In situations where cell culture facilities or expertise are not available, alternative methods for virus isolation can be employed. Clinical specimens can be inoculated intracranially into suckling mice or intrathoracically into mosquitoes. Suckling mice can develop encephalitis symptoms upon dengue infection, although some dengue strains may not cause overt illness in mice. In infected mice or mosquitoes, virus antigen can be detected in brain tissue or mosquito head squashes by staining with anti-dengue antibodies. These methods are less sensitive and specific compared to cell culture but can be useful in resource-limited settings.
3.2. Nucleic Acid Detection Methods
Nucleic acid detection methods, primarily targeting dengue viral RNA, have revolutionized dengue diagnosis due to their rapid turnaround time and high sensitivity. However, similar to virus isolation, RNA is also heat-labile, requiring careful specimen handling and storage to prevent degradation and ensure accurate test results. Specimens for nucleic acid detection must be collected, handled, and stored according to the same stringent procedures as those for virus isolation, emphasizing refrigeration or freezing to maintain RNA integrity.
3.2.1. Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Since the 1990s, reverse transcriptase-polymerase chain reaction (RT-PCR) assays have become a cornerstone of dengue diagnostics. RT-PCR offers significantly improved sensitivity compared to virus isolation and provides results much more rapidly, typically within hours instead of days or weeks. In situ RT-PCR, a specialized technique, allows for the detection of dengue RNA directly within paraffin-embedded tissue sections, which is particularly valuable for retrospective studies and understanding tissue-specific viral distribution.
All nucleic acid detection assays, including RT-PCR, involve three fundamental steps: 1) nucleic acid extraction and purification, 2) amplification of the nucleic acid target, and 3) detection and characterization of the amplified product. The initial step, RNA extraction and purification from clinical specimens, was traditionally performed using liquid phase separation methods, such as phenol-chloroform extraction. However, these methods have largely been replaced by silica-based commercial kits. These kits, available in bead or column formats, offer greater reproducibility, are faster, and are amenable to automation using robotics systems, improving efficiency and throughput in diagnostic laboratories.
Many laboratories utilize nested RT-PCR assays for dengue detection and serotyping. Nested RT-PCR involves two rounds of PCR amplification to enhance sensitivity and specificity. Typically, universal dengue primers targeting the C/prM region of the dengue genome are used in the first round of reverse transcription and amplification. This is followed by a nested PCR amplification step using serotype-specific primers to identify the specific dengue serotype (DENV-1, -2, -3, or -4) causing the infection.
An alternative approach is one-step multiplex RT-PCR. This method combines all four serotype-specific oligonucleotide primer sets in a single reaction tube. Multiplex RT-PCR simplifies the procedure and reduces the risk of contamination compared to nested RT-PCR. Following PCR amplification, the products are separated by electrophoresis on an agarose gel. The amplified DNA fragments are visualized as bands of different molecular weights in the agarose gel after staining with ethidium bromide dye. These bands are then compared with standard molecular weight markers to determine their size. In this assay design, dengue serotypes are identified based on the characteristic size of their amplified DNA bands.
The sensitivity of RT-PCR methods, compared to virus isolation, ranges from 80% to 100% in various studies. This variability in sensitivity depends on several factors, including the specific region of the dengue genome targeted by the primers, the RT-PCR assay format (e.g., one-step vs. two-step RT-PCR), and the method used for serotyping (e.g., nested PCR, blot hybridization with specific DNA probes, restriction site-specific PCR, sequence analysis, etc.). To minimize false-positive results due to non-specific amplification, it is crucial to design primers that target dengue-specific genome regions that are not conserved among other flaviviruses or related viruses. False-positive results can also arise from contamination by amplicons from previous PCR reactions. Stringent laboratory protocols and physical separation of different steps of the RT-PCR procedure, along with rigorous decontamination procedures, are essential to prevent contamination and ensure the reliability of RT-PCR results.
3.2.2. Real-Time RT-PCR Assays
Real-time RT-PCR assays represent a significant advancement over conventional RT-PCR. Real-time RT-PCR is a one-step assay system that allows for both the detection and quantification of viral RNA in real-time during the PCR reaction. This is achieved using primer pairs and fluorescent probes that are specific to each dengue serotype. The use of a fluorescent probe enables the detection of PCR reaction products as they are generated, in real-time, within a specialized PCR machine. This eliminates the need for post-PCR processing steps like electrophoresis, reducing assay turnaround time and the risk of contamination.
Numerous real-time RT-PCR assays have been developed using different fluorescence-based detection chemistries, primarily TaqMan and SYBR Green technologies. TaqMan real-time PCR is highly specific due to the sequence-specific hybridization of the fluorescent probe to the target DNA sequence. However, it’s important to note that the sensitivity of primers and probes reported in publications may vary, and they may not be able to detect all dengue virus strains equally well. The sensitivity is highly dependent on the homology between the primer and probe sequences and the targeted gene sequence of the specific virus being analyzed.
SYBR Green real-time RT-PCR offers the advantage of simplicity in primer design and utilizes universal RT-PCR protocols. SYBR Green is a fluorescent dye that binds to any double-stranded DNA, making primer design less constrained. However, SYBR Green-based assays are theoretically less specific than TaqMan assays because the dye can bind to any double-stranded DNA, including non-target PCR products.
Real-time RT-PCR assays can be designed as either “singleplex” or “multiplex.” Singleplex assays detect only one dengue serotype in each reaction, while multiplex assays are capable of simultaneously identifying all four dengue serotypes from a single sample. Multiplex assays offer the advantage of serotype identification in a single reaction, reducing assay time and minimizing the potential for contamination during sample handling. However, current multiplex real-time RT-PCR assays are often less sensitive compared to nested RT-PCR assays. A key advantage of real-time RT-PCR is its ability to determine the viral load or viral titer in a clinical sample. Viral load quantification can be valuable for studying the pathogenesis of dengue disease, monitoring disease progression, and assessing treatment response.
3.2.3. Isothermal Nucleic Acid Amplification Methods
Isothermal amplification methods offer alternatives to PCR-based nucleic acid detection by amplifying nucleic acids at a constant temperature, eliminating the need for thermal cycling instrumentation. Nucleic acid sequence-based amplification (NASBA) is one such isothermal RNA-specific amplification assay. In NASBA, the initial step is reverse transcription, where the single-stranded RNA target is copied into a double-stranded DNA molecule. This DNA molecule then serves as a template for RNA transcription, leading to RNA amplification. Detection of the amplified RNA can be achieved through electrochemiluminescence or in real-time using fluorescent-labeled molecular beacon probes. NASBA has been adapted for dengue virus detection and has shown sensitivity comparable to virus isolation in cell cultures. NASBA may be particularly useful for dengue diagnostic applications in field studies and resource-limited settings due to its isothermal nature and potential for rapid, point-of-care diagnostics.
Loop-mediated isothermal amplification (LAMP) methods have also been described for dengue virus detection. LAMP is another isothermal amplification technique known for its rapid amplification and ease of use. However, the performance characteristics of LAMP assays for dengue, in comparison to other nucleic acid amplification methods like RT-PCR and NASBA, are still being evaluated, and more data is needed to fully assess their diagnostic utility.
3.3. Antigen Detection Assays
The detection of dengue viral antigens in patient samples, particularly in acute-phase serum, provides a rapid and early diagnostic approach. Historically, antigen detection in secondary dengue infections was challenging due to the formation of virus-IgG antibody immunocomplexes, which could interfere with antigen detection assays. However, recent advancements in ELISA and dot blot assays targeting the envelope/membrane (E/M) antigen and the non-structural protein 1 (NS1) have demonstrated that high concentrations of these antigens, often in the form of immune complexes, can be detected in patients with both primary and secondary dengue infections, up to approximately nine days after the onset of illness.
The NS1 glycoprotein is produced by all flaviviruses and is secreted from infected mammalian cells. NS1 elicits a strong humoral immune response in infected individuals. The detection of NS1 antigen has emerged as a valuable tool for early dengue diagnosis. Several commercial kits for NS1 antigen detection are now widely available. These kits are generally rapid and easy to use, but most do not differentiate between dengue serotypes. The performance characteristics and clinical utility of these commercial NS1 antigen detection kits are currently being rigorously evaluated by laboratories worldwide, including through collaborative networks such as the WHO/TDR/PDVI laboratory network. These evaluations are crucial for determining the sensitivity, specificity, and overall diagnostic accuracy of NS1 antigen tests in various epidemiological settings and patient populations.
Beyond ELISA-based assays, fluorescent antibody assays, immunoperoxidase assays, and avidin-biotin enzyme assays can also be used for dengue virus antigen detection. These techniques are particularly useful for detecting dengue virus antigen in acetone-fixed leukocytes from patient blood samples and in snap-frozen or formalin-fixed tissues collected at autopsy. These methods can provide valuable information about viral distribution and tissue tropism in dengue infections.
3.4. Serological Diagnostic Tests
Serological tests are essential for dengue diagnosis, particularly after the acute viremic phase when antibody responses become detectable. Serological assays detect and measure antibodies produced by the host in response to dengue virus infection. The main types of antibodies measured are IgM, IgG, and IgA. Various serological assays are available, each with its own principles, advantages, and limitations.
3.4.1. IgM Antibody-Capture ELISA (MAC-ELISA)
The IgM antibody-capture enzyme-linked immunosorbent assay (MAC-ELISA) is a widely used serological test for dengue diagnosis, particularly for detecting recent dengue infections. In MAC-ELISA, total IgM antibodies present in the patient’s serum are captured by anti-μ chain-specific antibodies (specific to human IgM) that are pre-coated onto a microplate. Dengue-specific antigens, which can be derived from one to four dengue serotypes (DENV-1, -2, -3, and -4), are then added to the microplate. If dengue-specific IgM antibodies are present in the patient’s serum, they will bind to the captured IgM. These bound anti-dengue IgM antibodies are subsequently detected using monoclonal or polyclonal dengue antibodies that are directly or indirectly conjugated with an enzyme. This enzyme catalyzes a reaction that transforms a non-colored substrate into colored products. The intensity of the color, measured as optical density by a spectrophotometer, is proportional to the amount of dengue-specific IgM antibodies present in the sample.
Serum is the most common sample type for MAC-ELISA, but blood collected on filter paper and even saliva can also be used, although urine is not suitable. For IgM detection to be reliable, samples should be collected within the appropriate time frame, typically five days or more after the onset of fever, as IgM antibodies usually become detectable around this time. Serum specimens can be tested at a single dilution or at multiple dilutions to optimize assay sensitivity and dynamic range. Most antigens used in MAC-ELISA are derived from the dengue virus envelope protein (usually from virus-infected cell culture supernatants or suckling mouse brain preparations).
MAC-ELISA generally exhibits good sensitivity and specificity when used five or more days after fever onset. Numerous commercial MAC-ELISA kits and rapid diagnostic tests are available, but their sensitivity and specificity can vary significantly. A recent evaluation by the WHO/TDR/PDVI laboratory network of selected commercial ELISAs and first-generation rapid diagnostic tests found that ELISAs generally performed better than rapid tests in terms of diagnostic accuracy.
Cross-reactivity with other circulating flaviviruses, such as Japanese encephalitis virus, St. Louis encephalitis virus, and yellow fever virus, does not appear to be a major problem with MAC-ELISA. However, some false-positive results have been reported in sera from patients with malaria, leptospirosis, and past dengue infections. These limitations must be considered when using MAC-ELISA in regions where these other pathogens co-circulate with dengue. It is recommended that MAC-ELISA tests be evaluated against a panel of sera from relevant diseases prevalent in a particular region before widespread use in that area. It’s generally not possible to use IgM assays to identify specific dengue serotypes, as IgM antibodies tend to be broadly cross-reactive, even following primary dengue infections. However, some researchers have described MAC-ELISA modifications that may allow for serotype determination, but these require further evaluation and validation.
Alt text: Diagram illustrating the principle of a MAC-ELISA test, showing the steps of IgM capture, antigen binding, and enzyme-linked detection.
3.4.2. IgG ELISA
The IgG ELISA is another essential serological test used in dengue diagnostics. It is primarily used for detecting recent or past dengue infections. When paired sera (acute and convalescent samples) are collected within the correct time frame, IgG ELISA can help differentiate between recent and past infections. This assay typically uses the same dengue antigens as MAC-ELISA. A modification, the E/M-specific capture IgG ELISA (GAC), is designed to detect IgG antibodies that persist for an extended period, up to 10 months after infection. In contrast, indirect IgG ELISA assays coated with E/M antigen can detect IgG antibodies that persist for life, indicating past dengue exposure.
A four-fold or greater increase in IgG antibody levels between acute and convalescent paired sera, as measured by IgG ELISA, is a strong indicator of a recent dengue infection. The results of IgG ELISA generally correlate well with the haemagglutination-inhibition (HI) test, a traditional serological assay for flavivirus antibodies. An ELISA inhibition method (EIM) for detecting IgG dengue antibodies is also used for serological diagnosis and surveillance of dengue cases. EIM is based on the competition between IgG dengue antibodies in the patient sample and conjugated human IgG anti-dengue antibodies for binding to dengue antigen sites.
IgG ELISA can be used to detect IgG antibodies in serum, plasma, and blood samples stored on filter paper. It can also aid in differentiating between primary and secondary dengue infections. However, IgG ELISA generally lacks specificity within the flavivirus serocomplex groups. Antibodies produced shortly after a viral infection (acute phase) tend to have lower avidity (binding strength) compared to antibodies produced months or years after infection (convalescent phase).
Antibody avidity assays are used in some specialized laboratories to discriminate between primary and secondary dengue infections based on IgG avidity. However, these tests are not widely used in routine diagnostic settings and are not commercially available.
3.4.3. IgM/IgG Ratio
The IgM/IgG antibody ratio, specifically using dengue virus E/M protein-specific IgM and IgG, can be a useful parameter to distinguish between primary and secondary dengue virus infections. IgM capture and IgG capture ELISAs are the most common assays used for this purpose. In some laboratories, a dengue infection is classified as primary if the IgM/IgG optical density (OD) ratio is greater than 1.2 (using patient’s sera at a 1/100 dilution) or 1.4 (using patient’s sera at a 1/20 dilution). Conversely, an infection is considered secondary if the IgM/IgG ratio is less than 1.2 or 1.4, respectively. This algorithm for differentiating primary and secondary dengue infections based on IgM/IgG ratios has been adopted by some commercial vendors of dengue diagnostic kits. However, it’s important to note that these IgM/IgG ratio cutoffs can vary between laboratories, highlighting the need for better standardization of test performance and interpretation criteria across different diagnostic settings.
3.4.4. IgA Antibody Detection
Detection of serum anti-dengue IgA antibodies, measured by anti-dengue virus IgA capture ELISA (AAC-ELISA), is another serological approach explored in dengue diagnostics. IgA antibodies often become detectable in serum approximately one day after IgM antibodies appear. IgA antibody titers typically peak around day 8 after fever onset and then decline rapidly, becoming undetectable by around day 40 post-onset. Studies have not found significant differences in IgA titers between patients with primary and secondary dengue infections. Although IgA antibody levels are generally lower than IgM levels in both serum and saliva, some researchers suggest that performing both IgA and IgM assays together may improve the interpretation of dengue serology, particularly in complex cases. However, IgA antibody detection in dengue is not yet widely used in routine diagnostics and requires further evaluation to determine its clinical utility and added value.
3.4.5. Haemagglutination-Inhibition (HI) Test
The haemagglutination-inhibition (HI) test is a traditional serological assay that has been used for decades in flavivirus serology, including dengue. The HI test is based on the principle that dengue antigens can agglutinate red blood cells (RBCs) from ganders (male geese) or trypsinized human type O RBCs. Anti-dengue antibodies present in patient sera can inhibit this agglutination process. The potency of this inhibition is measured in the HI test, providing a measure of antibody titer.
Prior to performing the HI test, serum samples require pretreatment to remove non-specific inhibitors of haemagglutination. This is typically done by treating sera with acetone or kaolin. Sera are also adsorbed with gander or trypsinized type O human RBCs to remove non-specific agglutinins that might interfere with the assay. Each batch of dengue antigens and RBCs used in the HI test must be carefully optimized to ensure assay reliability and reproducibility. The pH optimum for haemagglutination varies for different dengue haemagglutinins, necessitating the use of multiple pH buffers for each dengue serotype to achieve optimal assay performance.
Ideally, the HI test requires paired sera: an acute-phase serum sample obtained upon hospital admission (or early in illness) and a convalescent-phase serum sample obtained at discharge (or at least seven days after the acute sample). The HI test does not discriminate between infections caused by closely related flaviviruses, such as dengue virus and Japanese encephalitis virus or West Nile virus. Furthermore, the HI test does not differentiate between immunoglobulin isotypes (IgM, IgG, etc.).
The antibody response in a primary dengue infection, as measured by the HI test, is characterized by low antibody levels in the acute-phase serum (drawn before day 5 of illness) and a slow, gradual increase in HI antibody titers thereafter. In contrast, during secondary dengue infections, HI antibody titers rise rapidly, typically exceeding 1:1280. Convalescent sera from patients with primary dengue responses generally exhibit HI antibody titers below this level.
Alt text: Diagram illustrating the principle of the Haemagglutination-inhibition assay, showing how antibodies inhibit the agglutination of red blood cells by dengue antigens.
3.5. Haematological Tests in Dengue Diagnosis
While not specific to dengue, haematological tests, particularly platelet counts and haematocrit values, are routinely measured during the acute stages of dengue infection and provide valuable clinical information for patient management. These tests should be performed carefully using standardized protocols, reagents, and equipment to ensure accurate and reliable results.
A decrease in the platelet count below 100,000 per microliter (μL) may be observed in dengue fever, but it is a consistent and prominent feature of dengue haemorrhagic fever (DHF). Thrombocytopaenia (low platelet count) typically occurs between day 3 and day 8 following the onset of illness. Monitoring platelet counts is crucial for assessing disease severity and guiding clinical management decisions, especially in dengue cases with haemorrhagic manifestations.
Haemoconcentration, indicated by an increase in haematocrit of 20% or more compared to convalescent values, is suggestive of hypovolaemia (decreased blood volume) due to increased vascular permeability and plasma leakage, a hallmark of severe dengue. Elevated haematocrit is a critical indicator of plasma leakage and impending circulatory compromise in dengue patients. Monitoring haematocrit levels, along with other clinical parameters, is essential for early recognition and management of severe dengue and to prevent progression to dengue shock syndrome.
4. Future Directions in Dengue Diagnostic Test Development
Advancements in diagnostic technologies are continuously shaping the landscape of dengue laboratory diagnosis. Several promising approaches are under development and evaluation, aiming to improve the speed, sensitivity, specificity, and accessibility of dengue diagnostic tests.
Microsphere-based immunoassays (MIAs) are gaining popularity as a versatile serological option for the laboratory diagnosis of numerous diseases, including dengue. MIA technology is based on the covalent bonding of antigens or antibodies to microspheres or beads. Detection of antibody-antigen interactions is typically achieved using lasers to elicit fluorescence of varying wavelengths from fluorescent dyes attached to the microspheres. MIA technology offers several advantages for dengue serology. It is generally faster than traditional MAC-ELISA and has significant potential for multiplexing serological tests. Multiplexing allows for the simultaneous detection of antibody responses to multiple dengue serotypes or even to different viruses in a single assay, increasing efficiency and reducing sample volume requirements. MIAs can also be adapted for virus detection, expanding their application beyond serology.
Rapid advances in biosensor technology, particularly those utilizing mass spectrometry, are leading to the development of powerful systems for rapid pathogen identification. Mass spectrometry-based biosensors can provide rapid discrimination of biological components in complex mixtures, such as patient samples. The mass spectra generated are unique to each pathogen and can be considered a specific “fingerprint” or molecular profile of the bacteria or virus analyzed. Sophisticated software systems integrated into these instruments can identify and quantify pathogens in a given sample by comparing the obtained mass spectra with extensive databases of infectious agents. This allows for the rapid identification of thousands of types of bacteria and viruses, including dengue viruses. Furthermore, these tools have the capability to recognize previously unidentified organisms in a sample and characterize their relationship to known pathogens. This could be invaluable for not only identifying dengue serotypes but also for characterizing dengue genotypes during outbreak investigations. Identification kits for infectious agents based on mass spectrometry are becoming available in 96-well format and can be customized to meet specific diagnostic requirements. The workflow typically involves sample processing for DNA extraction, PCR amplification, mass spectrometry analysis, and automated computer analysis for pathogen identification.
Microarray technology offers another promising avenue for multiplexed pathogen detection. Microarrays make it possible to screen a sample for a large number of different nucleic acid fragments simultaneously, corresponding to various viruses or pathogens. In the context of dengue, microarrays could be designed to detect and differentiate dengue virus serotypes and potentially other co-circulating arboviruses. Prior to hybridization to the microarray, the genetic material from the sample must be amplified. The amplification strategy can target conserved sequences common to all dengue serotypes or use random-based amplification to capture a broader range of viral sequences. Microarrays typically consist of short oligonucleotides or longer DNA fragments attached to a solid surface. Shorter oligonucleotides provide relatively exact sequence identification, while longer DNA fragments offer higher tolerance for mismatches, improving the ability to detect diverged strains or variants. A laser-based scanner is commonly used as a reader to detect amplified fragments that are labeled with fluorescent dyes. Microarray technology holds significant potential for comprehensive and multiplexed testing for dengue virus and other arboviruses circulating in a region, as well as for identifying all pathogens responsible for dengue-like symptoms in differential diagnosis.
Other innovative approaches are being explored in dengue diagnostics, although they are still in the early stages of development and evaluation. Luminescence-based techniques, for instance, are gaining interest due to their high sensitivity, low background signal, wide dynamic range, and relatively inexpensive instrumentation. Luminescence-based assays may offer advantages for dengue antigen and antibody detection, potentially leading to more sensitive and cost-effective diagnostic tests in the future.
5. Quality Assurance in Dengue Laboratory Diagnosis
Ensuring the quality and reliability of dengue laboratory diagnosis is of paramount importance for accurate patient care, effective disease surveillance, and valid research outcomes. Many laboratories, particularly in resource-limited settings, utilize in-house assays for dengue diagnosis. A significant challenge with in-house assays is the lack of standardization of protocols and reagents across different laboratories. This lack of standardization can lead to variability in test performance, making it difficult to compare results obtained from different laboratories or to aggregate data for surveillance purposes.
To address these challenges, it is crucial for national and regional reference centers to organize comprehensive quality assurance programs for dengue diagnostics. These programs should aim to ensure the proficiency of laboratory staff in performing dengue diagnostic assays and to provide reference materials for quality control of test kits and in-house assays. Proficiency testing, where laboratories are sent blinded samples to test and their results are evaluated against a reference standard, is an essential component of quality assurance programs. Reference materials, including well-characterized positive and negative control samples, are critical for internal quality control and for validating the performance of diagnostic assays.
For nucleic acid amplification assays, such as RT-PCR, stringent precautions are necessary to prevent contamination of patient materials and reagents. Contamination can lead to false-positive results and compromise diagnostic accuracy. Laboratories performing nucleic acid amplification assays must implement robust contamination control measures, including physical separation of pre- and post-amplification areas, unidirectional workflow, use of aerosol-barrier pipette tips, and regular decontamination of work surfaces and equipment. External quality assessment (EQA) schemes and proficiency testing are particularly important for nucleic acid amplification assays to ensure a high degree of confidence in the reliability and accuracy of test results.
6. Biosafety Considerations in Dengue Diagnostic Laboratories
The collection and processing of blood and other clinical specimens in dengue diagnostic laboratories pose a potential risk of exposure to infectious materials for healthcare workers and laboratory personnel. To minimize the risk of laboratory-acquired infections, adherence to safe laboratory techniques and biosafety practices is essential. These practices should be in accordance with established biosafety guidelines, such as those outlined in the World Health Organization’s Laboratory biosafety manual.
Key biosafety measures include the consistent use of personal protective equipment (PPE), such as gloves, laboratory coats, eye protection, and face masks, when handling patient specimens and infectious materials. Appropriate containers and procedures must be used for the safe collection, handling, and transport of specimens within and between laboratories. Proper waste management and decontamination procedures are critical for safely disposing of infectious waste and decontaminating laboratory surfaces and equipment. Laboratory personnel should receive comprehensive training on biosafety principles, safe laboratory practices, and emergency procedures to ensure a safe working environment and minimize the risk of accidental exposures to dengue virus and other pathogens.
7. Organization of Dengue Laboratory Services in Endemic Countries
In dengue-endemic countries, the effective organization of laboratory services is crucial for supporting patient care, public health surveillance, and outbreak response. Laboratory services should be strategically organized within the healthcare system to ensure accessibility, efficiency, and quality. Appropriate resources, including infrastructure, equipment, reagents, and trained personnel, must be allocated to support dengue diagnostic activities at different levels of the healthcare system. Training programs for laboratory staff are essential to build and maintain expertise in dengue diagnostic testing and quality assurance.
A tiered laboratory network model is often recommended for dengue-endemic countries. This model typically includes peripheral laboratories at primary healthcare facilities, intermediate-level laboratories at district or regional hospitals, and national or reference laboratories at the apex of the network. Peripheral laboratories may focus on rapid diagnostic tests for initial screening and case detection. Intermediate-level laboratories can perform a broader range of diagnostic tests, including ELISA-based assays and basic molecular tests. National or reference laboratories provide advanced diagnostic capabilities, including virus isolation, RT-PCR, serotyping, and specialized serological assays. They also serve as centers for quality assurance, training, and reference testing for the entire laboratory network. Effective coordination and communication within the laboratory network are essential for efficient sample referral, data sharing, and overall system performance.
Alt text: Table outlining a proposed model for organizing dengue laboratory services in endemic countries, suggesting tiered levels and corresponding diagnostic capabilities.
Alt text: Table providing examples of good and bad practices in dengue laboratory diagnosis, highlighting aspects like sample handling, test selection, and result interpretation.
By strategically organizing laboratory services, dengue-endemic countries can enhance their capacity for timely and accurate dengue diagnosis, strengthening both clinical care and public health response to this important infectious disease.
8. References
- Vorndam V, Kuno G. Gubler DJ, Kuno G. Dengue and dengue hemorrhagic fever. New York: CAB International; 1997. Laboratory diagnosis of dengue virus infections; pp. 313–333.
- Innis B, et al. An enzyme-linked immunosorbent assay to characterize dengue infections where dengue and Japanese encephalitis co-circulate. American Journal of Tropical Medicine and Hygiene. 1989;40:418–427. PubMed: 2540664
- PAHO. Dengue and dengue hemorrhagic fever in the Americas: guidelines for prevention and control. Washington, DC: Pan American Health Organization; 1994. (Scientific Publication No. 548)
- WHO. Dengue haemorrhagic fever: diagnosis, treatment, prevention and control. 2nd ed. Geneva: World Health Organization; 1997.
- Chanama S, et al. Analysis of specific IgM responses in secondary dengue virus infections: levels and positive rates in comparison with primary infections. Journal of Clinical Virology. 2004;31:185–189. PubMed: 15465410
- Kuno G, Gomez I, Gubler DJ. An ELISA procedure for the diagnosis of dengue infections. Journal of Virological Methods. 1991;33:101–113. PubMed: 1939502
- Shu PY, et al. Comparison of a capture immunoglobulin M (IgM) and IgG ELISA and non-structural protein NS1 serotype-specific IgG ELISA for differentiation of primary and secondary dengue virus infections. Clinical and Diagnostic Laboratory Immunology. 2003;10:622–630. PMC free article: PMC164246 PubMed: 12853395
- Falconar AK, de Plata E, Romero-Vivas CM. Altered enzyme-linked immunosorbent assay immunoglobulin M (IgM)/IgG optical density ratios can correctly classify all primary or secondary dengue virus infections 1 day after the onset of symptoms, when all of the viruses can be isolated. Clinical and Vaccine Immunology. 2006;13:1044–1051. PMC free article: PMC1563575 PubMed: 16960117
- Pelegrino JL. Summary of dengue diagnostic methods. World Health Organization, Special Programme for Research and Training in Tropical Diseases; 2006. (unpublished report)
- Hunsperger EA, et al. Evaluation of commercially available anti–dengue virus immunoglobulin M tests. Emerging Infectious Diseases. 2009. (serial online) March (date cited) Accessible at http://www.cdc.gov/EID/content/15/3/436.htm. PMC free article: PMC2681117 PubMed: 19239758
- Morens DM, et al. Simplified plaque reduction neutralization assay for dengue viruses by semimicro methods in BHK-21 cells: comparison of the BHK suspension test with standard plaque reduction neutralization. Journal of Clinical Microbiology. 1985;22(2):250–254. PMC free article: PMC268369 PubMed: 4031038
- Alvarez M, et al. Improved dengue virus plaque formation on BHK21 and LLCMK2 cells: evaluation of some factors. Dengue Bulletin. 2005;29:1–9.
- Halstead SB, Rojanasuphot S, Sangkawibha N. Original antigenic sin in dengue. American Journal of Tropical Medicine and Hygiene. 1983;32:154–156. PubMed: 6824120
- Lanciotti RS, et al. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. Journal of Clinical Microbiology. 1992;30:545–551. PMC free article: PMC265106 PubMed: 1372617
- Harris E, et al. Typing of dengue viruses in clinical specimens and mosquitoes by single tube multiplex reverse transcriptase PCR. Journal of Clinical Microbiology. 1998;36:2634–2639. PMC free article: PMC105176 PubMed: 9705406
- Vaughn DW, et al. Dengue viremia titer, antibody response pattern and virus serotype correlate with disease severity. Journal of Infectious Diseases. 2000;181:2–9. PubMed: 10608744
- Shu PY, Huang JH. Current advances in dengue diagnosis. Clinical and Diagnostic Laboratory Immunology. 2004;11(4):642–650. PMC free article: PMC440621 PubMed: 15242935
- Parida MM, et al. Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. Journal of Clinical Microbiology. 2005(43):2895–2903. (10.1128/JCM.43.6.2895-2903.2005) PMC free article: PMC1151941 PubMed: 15956414
- Vazquez S, et al. Serological markers during dengue 3 primary and secondary infections. Journal of Clinical Virology. 2005;33(2):132–137. PubMed: 15911428
- Fernandez RJ, Vazquez S. Serological diagnosis of dengue by an ELISA inhibition method (EIM). Memórias do Instituto Oswaldo Cruz. 1990;85(3):347–351. PubMed: 2134709
- Vazquez S, Fernandez R, Llorente C. Usefulness of blood specimens on paper strips for serologic studies with inhibition ELISA. Revista do Instituto de Medicina Tropical de São Paulo. 1991;33(4):309–311. PubMed: 1844953
- Vazquez S, et al. Kinetics of antibodies in sera, saliva, and urine samples from adult patients with primary or secondary dengue 3 virus infections. International Journal of Infectious Diseases. 2007;11:256–262. PubMed: 16914345
- Nawa M. Immunoglobulin A antibody responses in dengue patients: a useful marker for serodiagnosis of dengue virus infection. Clinical and Vaccine Immunology. 2005;12:1235–1237. PMC free article: PMC1247829 PubMed: 16210489
- Lemmer K, et al. External quality control assessments in PCR diagnostics of dengue virus infections. Journal of Clinical Virology. 2004;30:291–296. PubMed: 15163416
- WHO. Laboratory biosafety manual. 3rd ed. Geneva: World Health Organization; 2004. (ISBN 92 4 154650 6, WHO/CDS/CSR/LYO/2004.11 http://www.who.int/csr/resources/publications/biosafety/Biosafety7.pdf.