Background
In medicine, effective communication between doctors and patients is paramount for shared decision-making regarding treatment options and risk assessment. However, diagnostic errors are a persistent concern, with reported frequencies varying from 2.5% to as high as 20% [1, 2]. While many of these errors have minor consequences, some can be significantly detrimental to patient outcomes. The integration of modern computing technology holds promise for reducing diagnostic inaccuracies [3–5]. The advent of electronic history taking systems facilitates the collection of comprehensive patient data [5–16]. Our approach centers on the concept of patient data collection as the creation of a patient data vector. Assigning a diagnosis based on a patient’s symptoms and signs can be achieved through various methods: physician expertise, adherence to clinical pathways and guidelines, specialist boards, second opinions, and computer-assisted analysis of medical knowledge databases. The latter approach necessitates formalized systems capable of systematically sampling medical knowledge, specifically disease prototypes. This paper introduces a disease prototype database structured around the concept of vectors—patient vectors and disease vectors. This methodology requires three key elements: (1) a patient vector, (2) a repository of disease vectors (detailed herein), and (3) an algorithm to facilitate interaction between these vectors. Clinical decision support systems (CDSS) and inference engine technology are available to support approaches (2) and (3) [17–19]. Most existing computer-assisted medical knowledge databases rely on object-oriented structures and entity-relationship models. Further advancements in CDSS can be realized through IT systems that enhance knowledge acquisition, such as MetaMap [20] and cTake [21].
Our system demonstrates the feasibility of utilizing fundamental linguistic elements and an intuitive semantic network to capture medical context and structures for complex diagnostic understanding. The diagnostic process emerges as a system-driven dialogue aimed at aligning system semantics with patient semantics. This process can be conceptualized as a “linguistic game” that offers a “third opinion” on a patient’s symptoms, potentially improving diagnostic accuracy and acting as a valuable medical diagnosis support tool.
Methods
To illustrate our approach, we utilized four patient case examples, matching their symptoms to potential disease entities, which we term “pathosoms.” This Medical Diagnosis Game approach helps in structuring and analyzing complex medical information.
Examples
Case 1: A patient exhibiting cytopenia, anemia, and hypercellular bone marrow (3-tuple, one with an attribute).
Case 2: A 55-year-old female presenting with a firm lump in the breast (2-tuple with 1 and 2 attributes).
Case 3: A newborn infant with “polydactyly,” “renal cyst,” and “encephalocele” (3-tuple).
Case 4: A female patient experiencing proteinuria, hematuria, and fever (3-tuple).
Thesauri
A thesaurus, in our context, is defined as a collection of atomistic terms with a coding system (numeric, alpha-numeric, or string). Numerous medical thesauri are accessible online or through institutions like DIMDI (Deutsches Institut für medizinische Dokumentation) [22]. Examples include ICD (International Classification of Diseases, injuries and causes of death [23], ICD-O (International Classification of Diseases, Oncology) [24], ORPHANET (Portal für seltene Krankheiten und Orphan Drugs) [25], TA (Taxonomy of Anatomy) [26], OPS (Operationen-und Prozedurenschlüssel -Internationale Klassifikation der Prozeduren in der Medizin 2015) [27], OMIM (Online Mendelian Inheritance in Man) [28], CAS (common chemistry data base) [29], EC (comprehensive enzyme information system) [30], HGNC (Hugo Gene Nomenclature Committee 2015) [31], ATC (Anatomical Therapeutic Chemical Classification) [32] and LOINC (Logical Observation identifiers and names) [33]. Memem7 harmonizes and incorporates these thesauri terms and codes using our proprietary coding system, linked to the original codes.
Software
The system’s design emphasizes flexibility, utilizing common website programming environments such as SQL databases and Javascript. This ensures accessibility and ease of integration within existing healthcare IT infrastructures, making it a practical medical diagnosis game platform.
Atomistic Approach
Each symptom or sign descriptor (pathological sign, laboratory finding, EKG report, or genetic finding) is converted into an atomistic term. These atomistic terms form the foundational elements of our system. An atomistic term can be viewed as a base word with a unique code and associated attributes. For example, “proteinuria” with a numeric attribute (g/ml) or “headache” with the attribute “strong.” Attributes can be cardinal, ordinal, or numeric, allowing for nuanced descriptions within the medical diagnosis game.
Prototypic Diseases (Pathosom)
A disease entity or prototype, termed “pathosom,” is defined as the aggregate of all descriptors found in WHO classifications or clinical pathways. All descriptors must be atomistic, forming “pathophemes.” Images are integrated via internet links. These descriptors are categorized into classes as outlined in Table 1. A pathosom is thus represented as a vector sum: c (j1…jn), where each element ‘j’ belongs to specific classes (Table 1) or thesauri (Table 2).
Table 1. Classes of Memem7
| No | Class | Subclass | Subclass2 | Elements |
|---|---|---|---|---|
| 1100 | Description | Definition | 7241 | |
| 1400 | Description | System/Lokalisation | 9875 | |
| 1500 | Description | Struktur | ElementOf | 1733 |
| 1540 | Description | Struktur | HasElement | 6251 |
| 1550 | Description | Struktur | HasVariante | 1651 |
| 1570 | Description | Struktur | Sekundare Form | 177 |
| 5100 | Symptoms | Anamnese | 7406 | |
| 5110 | Symptoms | Anamnese | Akut | 182 |
| 5130 | Symptoms | Anamnese | Vorgeschichte | 479 |
| 5140 | Symptoms | Anamnese | Familie | 47 |
| 5150 | Symptoms | Anamnese | Demografisch | 10 |
| 5160 | Symptoms | Anamnese | Sozial | 88 |
| 5300 | Symptoms | Vital | 7597 | |
| 5400 | Symptoms | Physikal | 768 | |
| 5410 | Symptoms | Physikal | Spirometry | 22 |
| 5500 | Symptoms | Labor | 4423 | |
| 5510 | Symptoms | Labor | Clinical | 696 |
| 5520 | Symptoms | Labor | Agent | 1370 |
| 5530 | Symptoms | Labor | Toxicology | 35 |
| 5600 | Symptoms | Imaging | 401 | |
| 5610 | Symptoms | Imaging | Ultrasound | 119 |
| 5620 | Symptoms | Imaging | Radiology | 570 |
| 5630 | Symptoms | Imaging | MRT | 187 |
| 5640 | Symptoms | Imaging | CT | 132 |
| 5650 | Symptoms | Imaging | Endoskopie | 16 |
| 5700 | Symptoms | Pathologie | 1795 | |
| 5710 | Symptoms | Pathologie | Makroskopie | 1252 |
| 5720 | Symptoms | Pathologie | Mikroskopie | 8576 |
| 5725 | Symptoms | Pathologie | Elektronenmikroskopie | 291 |
| 5730 | Symptoms | Pathologie | Spezialfarbung | 236 |
| 5735 | Symptoms | Pathologie | Enzymhistochemie | 96 |
| 5738 | Symptoms | Pathologie | Zytologie | 894 |
| 5740 | Symptoms | Pathologie | Immunhistochemie | 3596 |
| 5745 | Symptoms | Pathologie | FACS | 29 |
| 5750 | Symptoms | Pathologie | In-Situ Hybridisierung | 39 |
| 5760 | Symptoms | Pathologie | Molekularbiologie | 30 |
| 5770 | Symptoms | Pathologie | Stoffe | 95 |
| 5795 | Symptoms | Pathologie | Differenzialdiagnose | 1356 |
| 5800 | Symptoms | Genetik | 4400 | |
| 5900 | Symptoms | Psychologie | 175 | |
| 6100 | Characteristics | Historie | 83 | |
| 6300 | Characteristics | Epidemologie | 2029 | |
| 6310 | Characteristics | Epidemologie | Sex | 557 |
| 6320 | Characteristics | Epidemologie | Age | 989 |
| 6330 | Characteristics | Epidemologie | Race | 66 |
| 6340 | Characteristics | Epidemologie | Region | 132 |
| 6350 | Characteristics | Epidemologie | Inzidenz | 198 |
| 6360 | Characteristics | Epidemologie | Pravalenz | 528 |
| 6400 | Characteristics | Atiologie | 607 | |
| 6500 | Characteristics | Pathophysiologie | 2783 | |
| 6600 | Characteristics | Verlauf | 588 | |
| 6610 | Characteristics | Verlauf | Beginn | 236 |
| 6620 | Characteristics | Verlauf | Verlauf | 266 |
| 6630 | Characteristics | Verlauf | Stadium | 462 |
| 6640 | Characteristics | Verlauf | Prognose | 1122 |
| 6650 | Characteristics | Verlauf | Komplikation | 1506 |
| 6660 | Characteristics | Verlauf | Risikofaktor | 965 |
| 6700 | Characteristics | Komorbiditat | 1316 | |
| 6800 | Characteristics | Differentialdiagnose | 7785 | |
| 6900 | Characteristics | Untersuchung | 3016 | |
| 8100 | Therapy | Therapieprinzipien | 1422 | |
| 8200 | Therapy | Medikamente | 2302 | |
| 8300 | Therapy | Chirurgie | 493 | |
| 8400 | Therapy | Strahlentherapie | 32 | |
| 8500 | Therapy | Ambulance | 8 | |
| 8600 | Therapy | ReHa | 44 | |
| 8700 | Therapy | Psychotherapie | 7 | |
| 8800 | Therapy | Alternative | 369 | |
| 9800 | Therapy | Vorsorge | 326 |
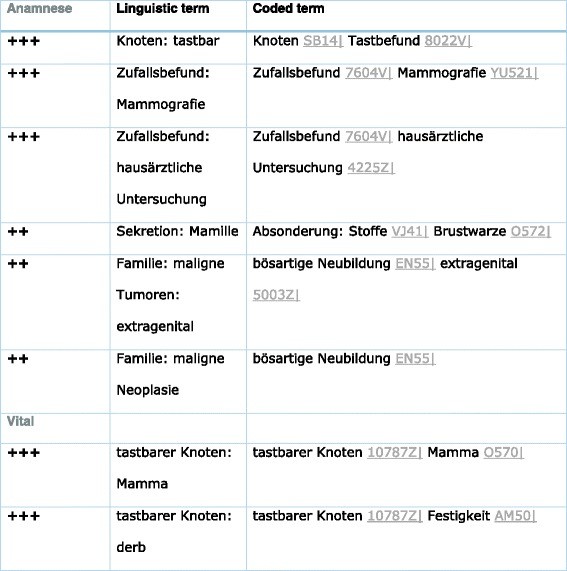
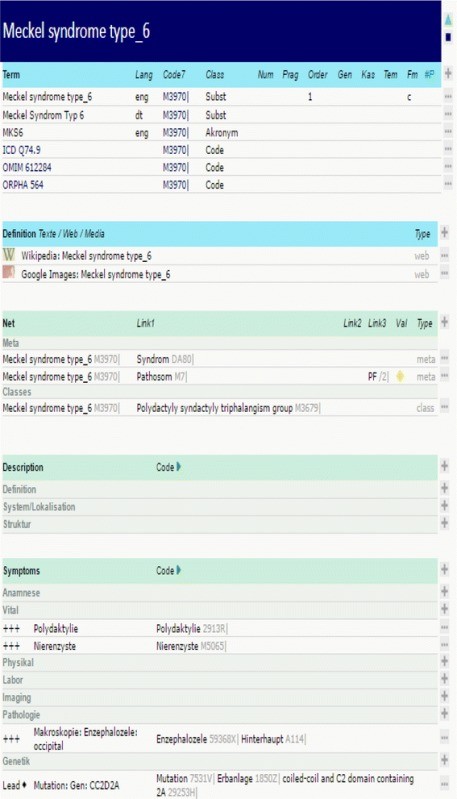
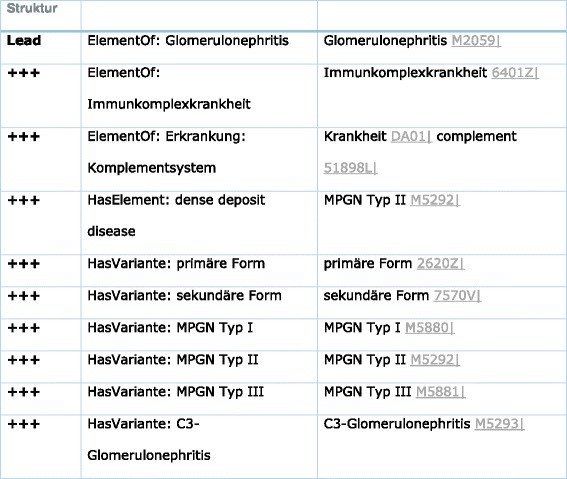
Table 2. Thesauri in medical use [21–29, 35, 36, 38]
| Thesaurus (acronym) | Content of diseases | Free available and used in memem7 |
|---|---|---|
| ICD-10-GM | Diseases | yes |
| ICD-O | Morphology of tumors | yes |
| ORPHANET | Rare Diseases | yes |
| OPS | Medical interventions | yes |
| LOINC | Laboratory Codings | yes |
| SNOMED | Medical terms and ontology | no |
| OMIM | Human genes/Genetic Disorders | yes |
| TA | Taxanomica anatomica | yes |
| CAS | Molecules | yes |
| EC | Enzymes | yes |
| HGNC | Molecular biological terms | yes |
| ATC | Agents/Medicaments | yes |
Pathophem
A “pathophem” is a structured sentence within a classification, clinical pathway, or publication, represented in its atomistic form. Following this breakdown, we have a core object (a single word representing a stem word linguistically), further defined by up to three attributes or objects. Attributes encompass time, intensity, color, taste, etc. Each object can be designated as true, false, or unavailable. Objects are coded using the thesaurus systems listed in Table 2. All content is coded within our system and termed “pathophem.” Additionally, occurrence probabilities ranging from 0 to 1 can be assigned. “Lead” indicates a pathophem invariably present in a pathosom (probability 1), “+++” signifies a frequency of 50% to 99.9% (probability 0.5–0.99), “++” denotes 10% to 49.9% (probability 0.1–0.49), “+” represents 1% to 9.9% (probability 0.01–0.09), and “(+)” represents frequencies below 1%. This probabilistic approach adds a layer of sophistication to the medical diagnosis game.
Diagnostic Algorithm (Linguistic Game)
The diagnostic algorithm employs a range of syntactical and semantic analysis techniques to generate a set of potential diagnoses. Diagnosis proposals arise from matching a patient vector (in our test system, a 5-tuple vector of patient data representing a medical report) with disease vectors (pathosoms). Initial analysis involves syntactical verification of the patient vector, ensuring all terms are present within the core thesauri and possess a “primary meaning” within the semantic network. Our semantic network is a conventional knowledge representation network, a directed graph with vertices representing terms, objects, and concepts, and edges denoting semantic relationships such as “is-a,” “is-part-of,” “has-attribute-of,” and “is-class-of.”
A continuous dialogue is established, offering alternative interpretations if terms are not fully matched, utilizing synonyms, clarifying ambiguous meanings, etc. The initial output is a ranked set of matching pathosoms. If this set is excessively large, the system conducts a secondary analysis to refine the set by requesting further details on essential pathosom symptoms. Conversely, if the set is very small or empty, the system broadens the patient vector using semantic network context methods like “is-class” and others. For instance, if a specific finger symptom/pathophem is sought but not found, the system may suggest a related hand symptom/pathophem that could be relevant. This interactive and iterative process embodies the “linguistic game” aspect of our medical diagnosis system.
Linguistic Problems (German/English)
Over 85% of the terms utilized are already translated into English, facilitating international applicability and future development for English-speaking medical professionals.
Statistical Methods
Statistical calculations were performed using the R statistic package (version 3.1.1). For ICD-10 (German version) analysis, the tm package of R [34] was employed to determine the number of atomized terms within ICD-10 and integrate any missing terms into Memem7, continuously improving our medical knowledge base.
Description of Software
- Project name: Memem7 Medical Semantic Network
- Project home page: not yet determined
- Archived version: not yet determined
- Operating system(s): Platform independent
- Programming language: Coldfusion, Javascript, HTML5
- Other requirements: Coldfusion Server
- License: Not yet determined
- Any restrictions to use by non-academics: Not yet determined
Evaluation of Memem7
Memem7’s quality was assessed using 190 artificial reports (simulated cases, not actual patient data), initially developed for testing CLEOS [3–5]. Each case comprised a brief medical report or symptoms with a mono-causal medical explanation, providing a robust test set for our medical diagnosis game.
Results
Examples for Using Memem7 for Searching
This involves comparing the patient vector (a tuple of up to 5 elements) with the pathosom set to identify pathosoms that best match the patient vector using a linguistic approach. This exemplifies the practical application of our medical diagnosis game.
Case 1
Search terms in English: cytopenia (41), anemia (657), bone marrow (91): hypercellular. This is a 3-element patient vector. Numbers in parentheses indicate pathophem mentions in Memem7. The intersection of these three pathophems yielded only one pathosom: RAEB (M3501, see Additional file 1).
Case 2
Search terms: female (597): aged 55 years, nodule (365): firm (90), nodule: mamma (434). Memem7 identified invasive breast cancer as the primary pathosom. Excluding the “firm” attribute, it proposed 6 additional pathosoms: angiosarcoma of breast, secretory breast cancer, benign breast tumor, pseudoangiomatoid stroma hyperplasia, fibroadenoma, and breast papilloma.
Case 3
Search terms: polydactyly (40), renal cyst (14), encephalocele (6) in a newborn infant. Three pathosoms aligned with these terms: Meckel syndrome type 1, 3, 6.
Case 4
Search terms: proteinuria (61) and hematuria (72). Memem7 returned 25 pathosoms as potential disease vectors. Adding “fever” (622) narrowed the results to 8 additional pathosoms: cryoglobulinemia with vasculitis, periarteriitis nodosa, microscopic polyangiitis, hanta virus infection, hemorrhagic fever with renal syndrome, emphysematous pyelonephritis, infective glomerulonephritis, and acute interstitial nephritis.
Examples of Pathosoms Fitting Cases 1–4
Case 1: RAEB (Refractory Anemia with Excess of Blasts)
RAEB, a myelodysplasia subset, exemplifies a disease entity. The WHO classification of tumors of hematopoietic and lymphoid tissue [35] describes it over two pages (pp100–101, [22]). ICD-10-GM code: D46.2; ICD-O-coding: 9983/3. The description begins: “refractory anemia with excess blasts is a myelodysplastic syndrome (MDS) with 5–19% myeloblasts in the bone marrow (BM) or 2–19% blasts in the peripheral blood.” In Memem7, this translates to a vector of atomistic terms: c (refractory anemia, myeloblasts: bone marrow > 5%: AND: myeloblasts: bone marrow 2%). The entire Boolean term must be TRUE. This illustrates pathopheme generation in the RAEB pathosom. Coded version: M3094|; (O812| & 10209|: 5..20%) | (10221 T| &10209 T|: 2..20%). Additional file 1: Tables S1 to S3 provide further details.
Case 2: Invasive Breast Cancer
Invasive breast cancer encompasses a range of clinical and morphological diseases with a hierarchical structure. There are 68 breast cancer pathosoms, including non-invasive and inflammatory breast cancer (Additional file 1: Tables S5 and S6), and tubular carcinoma, structured in a tree (“is-element,” “has-element,” “has-variant”). Inflammatory breast cancer, one of these 68 pathosoms, is detailed in Table 3 and Additional file 1: Tables S5 and S6.
Table 3. Part of the Pathosom Breast Cancer Concerning Anamnesis and Physical Examination (Vital)
Note that “tastbarer Knoten” (palpable node) is coded as 10787Z, encompassing lump and palpable node.
Case 3: Genetic Disorder of Skeletal Disease
Meckel syndrome type IV (Table 4) [36, 37] was used as an example. Atomistic terms are coded, and in cases of multiple codes from different thesauri, assignment is random. Table 1 shows six synonyms or codes, such as OMIM code 612284, facilitating further information retrieval within our medical diagnosis game framework.
Table 4. Meckel Syndrome VI as an Example of a Pathosom
Case 4: Membrano-Proliferative Glomerulonephritis (MPGN) (Table 5)
Table 5. Each Pathosom is Part of a Hierarchical Tree with Supersets and Subsets
Syntactic and language variations are irrelevant as “Nierenzyste,” “Nierencyste,” “renal cyst,” and “renal cysts” are all coded as M5065 within the system, highlighting the robustness of our medical diagnosis game across linguistic differences.
MPGN classification includes three subsets: MPGN Type I, II, and III [38]. ICD coding encompasses N05.5, N04.5, N03.5, N02.5, and N01.5 (Additional file 1: Tables S7 and S8).
Description of Memem7
Thesauri
Table 2 lists the thesauri in Memem7. For RAEB, the ICD-10 code is D46.2. RAEB is absent in other listed thesauri, but ICD-O [23] mentions “refractory cytopenia with multilineage dysplasia.” For genetic skeletal disorders, ICD-10 codes range from Q65.–Q79.9. Using R’s tm text analysis, we identified 53,947 atomized terms from ICD-10. 16,798 (31.1%) ICD-10 terms were initially missing in Memem7 and have been integrated. “Mallet finger” (German: Hammerfinger), coded as ICD-10 M20.0, was added after tm identified “Hammerfinger” in ICD-10. The ICD-10 diagnosis count (excluding signs, symptoms) is approximately 9700.
Number of Pathosoms and Pathophemes
Currently, Memem7 includes about 4600 pathosoms (prototypic diseases) with 104,200 pathophemes (prototypic symptoms). Terms from various thesauri (Table 2) are used for atomized pathophemes. Total coded terms are approximately 230,000 out of 1,550,000 terms across German, English, and Latin in different grammatical forms. Memem7 covers ~47.2% (4600/9700) of ICD-10 diagnoses. Each pathosom contains roughly 10–100 pathophemes, depending on disease complexity, demonstrating the depth of information within our medical diagnosis game.
Comparison of Pathosom and Diagnosis
A pathosom is a structured, flexible, non-deterministic data model of disease knowledge. It can be seen as a pathophem vector, with each pathophem being TRUE (1), FALSE (0), or having evidence between 0 and 1. A diagnosis is either an alpha-numeric code (e.g., ICD-10) [23, 24, 35] or a written disease description (e.g., RAEB) [35]. RAEB has variants like RAEB-1, RAEB-2, and RAEB-F, which are subsets of the RAEB pathosom, showcasing the hierarchical structure of our medical diagnosis game. RAEB, a 75-element vector (Additional file 1: Tables S1 to S4), is a subset of myelodysplasia, illustrating the “has-attribute” relationships within the semantic network’s tree structure.
Evaluation of the System
In 90 of 190 (47.4%) artificial cases, Memem7 correctly identified the proposed diagnosis. The number of proposed pathosoms (diagnoses) ranged from 0 to 173, with a median of 3, indicating a targeted and relevant output from our medical diagnosis game.
Discussion
Computational linguistics and computer technology are widely recognized as transformative forces in medical practice [39]. However, concrete evidence of improved medical quality from these changes is still emerging [18].
This paper details a software system comprising a pathosom pool (disease entities, syndromes, symptom sets). Our aim is to demonstrate the feasibility of computer-assisted symptom-to-disease entity assignment (using a preliminary 5n-tuple system). Prerequisites include: (1) atomistic linguistic knowledge delivery, (2) pathosoms as vectors, and (3) a linguistic game-based analysis assisting patient vector classification to pathosoms, without deterministic mathematical methods or distance measures. Memem7’s search function provides a ranked list of pathosoms explaining patient complaints, emphasizing linguistic and semantic network heuristics.
Future medical decision-making assistance systems require three tools: (1) electronic history taking, convertible to a patient data vector [3–5] or EHR extracts; (2) disease vectors (pathosoms) or disease entity algorithms; (3) an algorithm comparing patient and pathosom vectors, ranking fitting pathosoms (inference engine).
Memem7 currently enables: (1) pathosom vector construction and (2) limited dialogue with a 5n-tuple patient vector. “Limited dialogue” refers to the current 5-tuple analysis capacity. Post-dialogue, users receive n pathosoms matching the patient vector. Our approach’s primary goal was to prove the feasibility of formalized knowledge collection, creating a functional medical diagnosis game.
Mathematical methods from big data analysis are crucial for enhancing machine-assisted medical treatment decisions [40–42]. Memem7 covers approximately 4600 diagnoses, with ongoing expansion. Numerous electronic history-taking trials exist [43] (70+ by 2010) ([3, 4], review [5, 16]), but few open-source applications like OpenEMR and OpenEHR [44, 45] are available. In our linguistic game, these history-taking systems are replaced by a 5n-tuple. The “linguistic game” concept implies: (1) error is inherent, (2) decisions are assisted, not replaced, (3) user knowledge improves results (pathosom disease vectors). Thus, Memem7 acts as a third opinion for a 5-tuple patient vector. Integrating Memem7 with medical text recognition tools like MetaMap [20] or cTake [21] is underway, aiming to enhance the medical diagnosis game experience.
Regarding Memem7 accuracy, initial 5-tuple search term tests correctly identified the proposed diagnosis in 46.7% of 190 artificial cases. Clinical decision support system (CDSS) sensitivity data is scarce and inconsistent. Müller et al. [46] reported 96% sensitivity using New England Journal of Medicine case reports for systems like DXplain, Isabel Healthcare, Diagnosis Pro, PEPID [47–50]. Our lower initial accuracy is expected to improve with continuous testing and error correction, part of a quality improvement cycle enhancing both the semantic network and pathophemes.
Memem7 differs from systems like HELP [18] through its atomized linguistic term approach, contrasting with probabilistic, Bayesian, or machine learning systems. Combining linguistic and probabilistic methods is a future direction. Linguistic systems excel in easily adding atomized terms (pathophemes) to disease entities. Comparative analysis of linguistic and probabilistic approaches, or their synergy, is planned for the next software iteration, further refining our medical diagnosis game.
Many biomedical text mining and concept recognition tools exist [51, 52], focusing on natural language processing and concept identification via dictionary matching and machine learning. Memem7 uniquely focuses on using these concepts to identify complex entities like health disorders, rather than just concept identification. Future versions may handle concept identification from medical patient reports. Technically, Memem7 addresses ambiguous fact concept matching (symptoms, events, lab data) with complex concept trees (anatomy, body functions, diseases). The heuristic algorithms and concept plurivalence make the matching process more of a “game” than a deterministic process, hence “medical diagnosis game.”
Conclusions
In conclusion, we present an electronic disease entity system enabling formalized dialogue with patient data vectors, demonstrating a proof of concept for a linguistic game-based medical navigation system. This innovative medical diagnosis game holds promise for enhancing diagnostic processes.
Acknowledgements
We appreciate the Robert Bosch Foundation’s sustained support for medical practice improvement.
Funding
Supported by the Robert Bosch Foundation Stuttgart, Germany.
Availability of Data and Materials
Data and materials are available in a proof-of-concept state with limited test environment access. Datasets are available from the corresponding author upon reasonable request.
Authors’ Contributions
PF: medical knowledge input, conceptual work, system testing, manuscript preparation. AK: software development, conceptual work, manuscript preparation. PA: software development, conceptual work. FK: orphan disease input. CF: system testing, medical knowledge input. MDA: conceptual work, manuscript preparation. All authors contributed to manuscript reading and provided intellectual input throughout preparation. All authors approved the final manuscript.
Competing Interests
Dr. Peter Fritz: No competing interest.
Dr. Andreas Kleinhans: No competing interest.
Dipl-Inf. Patrick Albu: No competing interest.
Florian Kuisle: No competing interest.
Dr. Christine Fritz-Kuisle: No competing interest.
Prof. Mark Dominik Alscher: No competing interest.
Consent for Publication
Not applicable.
Ethics Approval and Consent to Participate
No patient data included; ethical approval not required per laws. Artificial patients are textbook-based. Baden-Wuerttemberg ethical board bylaws apply (“Gesetz über das Berufsrecht und die Kammern der Ärzte, Zahnärzte, Tierärzte, Apotheker, Psychologischen Psychotherapeuten sowie der Kinder- und Jugendlichenpsychotherapeuten (Heilberufe-Kammergesetz – HBKG) in der Fassung vom 16. März 1995 (GBl. BW v. 17. Mai 1995 S. 314)”).
Publisher’s Note
Springer Nature remains neutral regarding jurisdictional claims in published maps and institutional affiliations.
Abbreviations
ATC Anatomical therapeutic chemical classification
CAS Common chemistry data base
CDSS Clinical decision support system
DIMDI Deutsches Institut für medizinische Dokumentation
EC Brenda, comprehensive enzyme information system
HGNC Hugo gene nomenclature committee 2015)
HTML5 Hypertext markup language
ICD-10 International classification of diseases, injuries and causes of death
ICD-O International classification of diseases, oncology
LOINC Logical observation identifiers and names
MPNG Membrano-proliferative glomerulonephritis
OMIM Online Mendelian Inheritance in Man
OPS Operationen-und Prozedurenschlüssel-Internationale Klassifikation der Prozeduren in der Medizin 2015
ORPHANET Portal für seltene Krankheiten und Orphan Drugs
RAEB Refractory anaemia with excess blasts
TA Taxonomy of anatomy
WHO World Health Organization
Additional file
Additional file 1: Table S1. (168.6KB, docx) RAEB part 1. Table S2. RAEB part 2. Table S3. RAEB part 3. Table S4. RAEB part 4. Note that the underscored pathophem (IPSS Score) is not yet included in the system. Table S5. Inflammatory breast cancer part 1. Table S6. Inflammatory cancer part 2. Table S7. Membrano-proliferative glomerulonephritis part 1. Table S8. membrano-proliferative glomerulonephritis part 2. (DOCX 174 kb)
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12911-017-0488-3) contains supplementary material, available to authorized users.
Contributor Information
Peter Fritz, Email: [email protected].
Andreas Kleinhans, Email: [email protected].
Florian Kuisle, Email: [email protected].
Patricius Albu, Email: [email protected].
Christine Fritz-Kuisle, Email: [email protected].
Mark Dominik Alscher, Phone: +49-711-8101-3496, Phone: +49- 0711-8101-3792, Email: [email protected].
References
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Availability of data and materials: the system is in a proof-of-concept state and provides a limited test environment access. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.